creating soft links to images....
diamond>
ln -s /work/schreurs/diamond/testDataEVAL15/*.img.Z .
proteinsetup started on bekken at 25-Nov-2010 19:17:03 Created evpy-config Created view.init Created dirax.initCreate preliminary setup files for proteins.
Dirax volume test = voltest 112500.0 1125000.0 Created peakref.init proteinsetup ended at 25-Nov-2010 19:17:03 CPU time used 00:00:00
renameimages started on bekken at 25-Nov-2010 19:17:03 a search on *_*.img* revealed thaumatin_die_M1S5_1_0001.img.Z filetype=[img] img images are created by adsc, marccd and oxford detectors.The images in this case are named thaumatin_die_M1S5_1_0001.img.Z. Simple systematic names are easier to use. We have choosen for sxxfyyyy, where s stands for scan and f for frame. The first frame of the first scan will always be called s01f0001
Also, files of type img can originate from various
detectortypes (adsc, marccd and oxford). The filetype will also be
changed.
Enter detectortype (adsc/marccd/oxford) [adsc]
adsc
new filetype=[img] scanname=thaumatin_die_M1S5_ scannumber=1 framenumber=1 total 369 images (a,i2.2,'f',i4.4)new scan name [s] s
Created renameimages.log new scanname s01f renameimages ended at 25-Nov-2010 19:17:06 CPU time used 00:00:00
scancheckadsc started on bekken at 25-Nov-2010 19:17:06 ImageFileType=img Available scan names: s01f 369 images s01f0001.img.ZRead all images, and do some consistency checks.
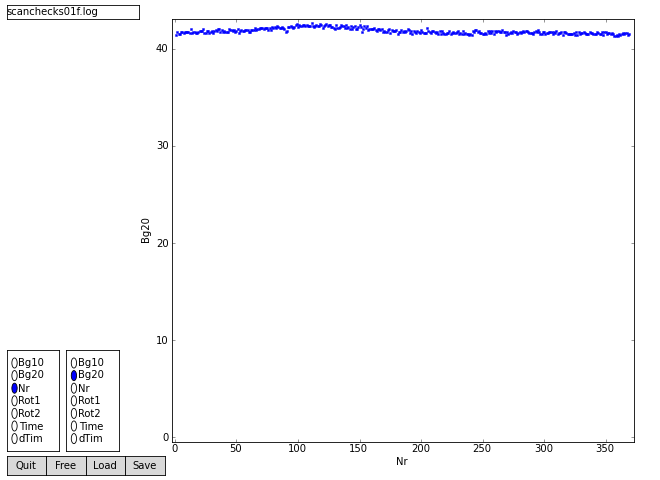
trying [s01f] s01f0001.img.Z Total 9437184 pixels stephor=10 stepver=10 Scanning 94249 pixels 369 images Time dTim Rot1 Rot2 Bg20 Bg10 FullTime Name 0.000 0.000 0.000 0.500 41.39 39.69 16:27:51.000 s01f0001.img.Z 3.000 3.000 0.500 1.000 41.72 40.01 16:27:54.000 s01f0002.img.Z 6.000 3.000 1.000 1.500 41.52 39.82 16:27:57.000 s01f0003.img.Z 8.000 2.000 1.500 2.000 41.54 39.85 16:27:59.000 s01f0004.img.Z 10.000 2.000 2.000 2.500 41.77 40.08 16:28:01.000 s01f0005.img.Z 12.000 2.000 2.500 3.000 41.73 40.04 16:28:03.000 s01f0006.img.Z ....cut.... 823.000 2.000 183.000 183.500 41.59 39.89 16:41:34.000 s01f0367.img.Z 825.000 2.000 183.500 184.000 41.40 39.73 16:41:36.000 s01f0368.img.Z 828.000 3.000 184.000 184.500 41.47 39.77 16:41:39.000 s01f0369.img.Z Created scanchecks01f.log Closed scanchecks01f.log one scancheck performed scancheck ended at 25-Nov-2010 19:20:38 CPU time used 00:02:41
['scanchecks01f.log'] 369 records from scanchecks01f.log radio: variables= ['Bg10', 'Bg20', 'Nr', 'Rot1', 'Rot2', 'Time', 'dTim'] 369 records from scanchecks01f.log horvar=Nr vervar=Bg20
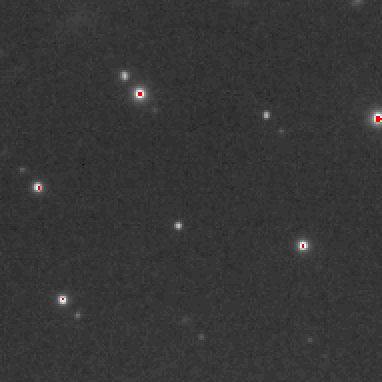
viewadscgd (version 1.4 2010101300) started on bekken at 25-Nov-2010 19:20:41Preliminary inspection of the first image
one frame from s01f0001.img.Z.
Lambda changed from 0.71073 to 1.488
Initialized DISTOR adsc920 Created detalign.vic 16 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0
Dist Swing Omega Chi Phi Axis Start End
old 0.00 0.00 0.00 0.00 0.00 1
new 116.00 0.00 0.00 0.00 0.00 3 0.00 0.50
The detector position (based on the offset numbers in the image header)
is written to detalign.vic.
Setting ZoomWindow 382 382
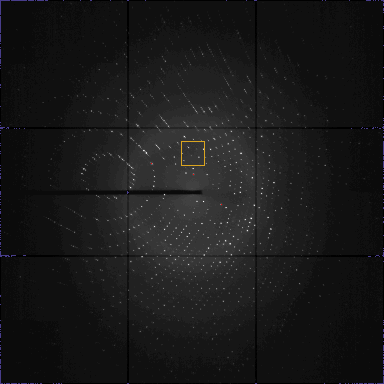
The initial detector offsets are read from the image header. Sometimes
these numbers are modified by the operator before data collection.
But what these numbers express is not documented (horizontal/vertical
or vertical/horizontal, from left to right or right to left, pointing up
or down). And if the operator did not update the information,
the header can contain outdated offsets.
The interpretation of the offsets from the header is in this case wrong.
The indexing is very sensitive for the position of the
primary beam. I am going to correct this here and now:
View>
detectoroffset
Current: 0.0 0.41 -0.11distance[0.0] <Enter>
Note on the syntax. To accept a proposed value, you may type Enter.
Commands and arguments may also be specified on one line. To accept a proposed
value in that construct, you may use a = (equal sign) or . (dot).
So the previous command could also be given by:
View>
detectoroffset = -0.11 0.41
Save this new offset. Later invocations of view will automatically
load this file.
View>
save detalign
WARNING: renaming existing file detalign.vic into detalign.vic.~1~ Created detalign.vic
Set the beamstop parameters. These are manually determined by
inspection of an image.
View>
beamstopwidth 4
View>
beamstopdiameter 0.1
View>
beamstop 8.5 0.2
Save the beamstop definition. Later invocations of view will automatically
load this file.
View>
save beamstop
Created beamstop.vic Closed beamstop.vic
What are the resolution limits?
View>
resomin
min resolution=28.42 (theta=1.5)Reso Min[28.42] <Enter>
min resolution=28.42 (theta=1.5)accept to proposed default.
Resolution half: Left 1.65 Right 1.65 Bottom 1.65 Top 1.65 max resolution=1.65 (theta=26.86)So the edges of the detector correspond to 1.65 Å (Memorize this number for later).
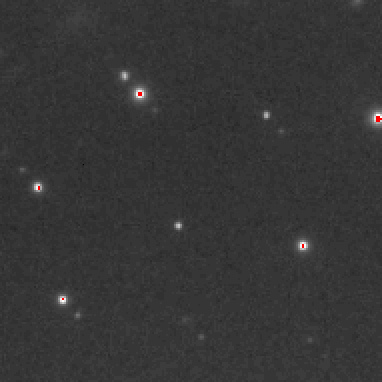
Do an initial peaksearch
View>
peak2
BadMaskFill 63181 pixels behind beamstop Tried=5 peaklist:5
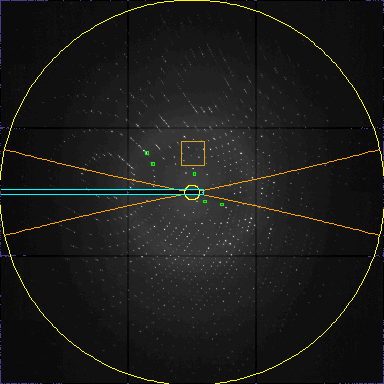
Set resolution limits to suitable values for indexing
View>
resomin 40
min resolution=40.0 (theta=1.07)View> resomax 3
max resolution=3.0 (theta=14.36)Increase the number of expected peaks
Tried=269 Duration=19 Theta=44 Respons=6 peaklist:200

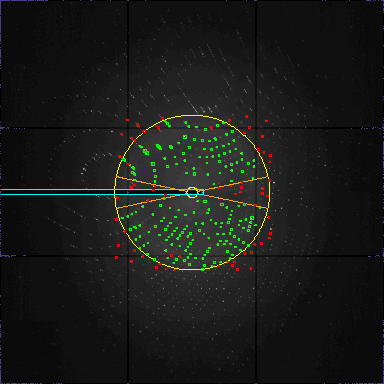
The dimension of the box around the peaks is set by peaksizemm. The
current value seems a bit large, with respect to the distance between
peaks. What is the current value?
View>
peaksizemm
peak size mm[1.2]
<Enter>
1.2 is a bit too large. Probably 0.8 is better.
View>
exit
view ended at 25-Nov-2010 19:20:44 CPU time used 00:00:03
buildsearch started on bekken at 25-Nov-2010 19:20:45 Found Z compressed .img images Available scans: scan s01f 369 images first image=s01f0001.img.Z Total 369 imagesprefix name [i] i
search types: 2: peak search in every frame. The total number of peaks is the number of expected peaks per frame times the number of frames. 3: peak search in first frame. peaksearch in second frame. Select the peaks in the previous frame, if they were stronger. Continue this process for n frames. The total number of peaks will be less than the number of expected peaks per frame times the number of frames. 4: repeat searchtype 3 over many images.search type (2/3/4) [3] 3
Created search3.vic Created isearch.vicEnter name of first image s01f0001
s01f0001.img.Z (s01f0001.img.Z exists) 369 image files nsearch 8 repcount 7more [No] no
buildsearch ended at 25-Nov-2010 19:20:45 CPU time used 00:00:00
viewadscgd (version 1.4 2010101300) started on bekken at 25-Nov-2010 19:20:45Run the script created with buildsearch
! Created by buildsearch at 25-Nov-2010 19:20:45
! Host bekken User schreurs WD /work/schreurs/diamond/diamond/
evpylog searching peaks type 3
! abort on warnings
abort on
! check contiguous images
checknext on
! set number of peaks in one image
peaknr 200
! peak quality
peakbg 3.0
peakbg2 3.0
! forbidden area
durationmax 5.0
! minimum intensity
peakmin 5
! no peaks to be found in next x degrees
peakremove 3.0
! stop after successive weaks
peakbreak 5
! peak size parameters
peaksizemm 0.8
! resolution limits
resomin 40.0
min resolution=40.0 (theta=0.51)
resomax 3.0
max resolution=3.0 (theta=6.8)
! apply new detalign if it exists
&detalign_i
! display n consecutive images
imageon 3
! do not display n intermediate images
imageoff 1000
! try read dark image later
trydark on 1.0
@search3(s01f0001.img.Z;i1;7)
one frame from s01f0001.img.Z.
Lambda changed from 0.71073 to 1.488 thmin changed to 1.07 thmax changed to 14.36
Initialized DISTOR adsc920 16 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0
Dist Swing Omega Chi Phi Axis Start End
old 0.00 0.00 0.00 0.00 0.00 1
new 116.00 0.00 0.00 0.00 0.00 3 0.00 0.50
Setting ZoomWindow 382 382
Created i1.drx
Created i1.pk
BadMaskFill 63181 pixels behind beamstop
Tried=267 Duration=19 Theta=43 Respons=5
peaklist:200 peakfile:0 diraxfile:0

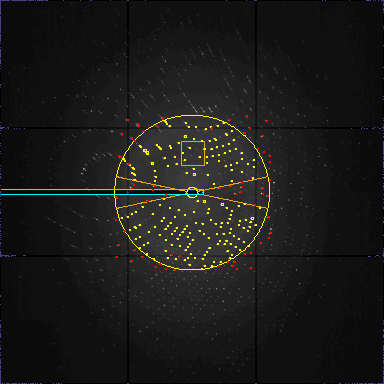
one frame from s01f0002.img.Z. 60 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0 200 peaks in list: OnFirstFrame=165 Improving=35 Tried=270 Duration=23 Theta=41 Respons=6 peaklist:200 peakfile:0 diraxfile:0

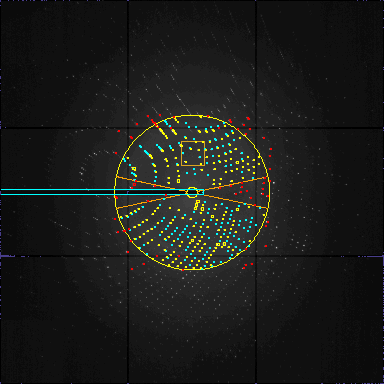
one frame from s01f0003.img.Z. 44 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0 200 peaks in list: Improving=35 Ok=165 Tried=264 Duration=18 Theta=41 Respons=5 peaklist:200 peakfile:165 diraxfile:165
one frame from s01f0004.img.Z. 54 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0 200 peaks in list: Improving=36 Ok=164 Tried=267 Duration=21 Theta=42 Respons=4 peaklist:200 peakfile:329 diraxfile:329 one frame from s01f0005.img.Z. 37 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0 200 peaks in list: Improving=44 Ok=156 Tried=277 Duration=25 Theta=45 Respons=7 peaklist:200 peakfile:485 diraxfile:485 one frame from s01f0006.img.Z. 30 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0 200 peaks in list: Improving=51 Ok=149 Tried=253 Duration=20 Theta=28 Respons=5 peaklist:200 peakfile:634 diraxfile:634 one frame from s01f0007.img.Z. 52 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0 200 peaks in list: Improving=33 Ok=167 Tried=280 Duration=19 Theta=53 Respons=8 peaklist:200 peakfile:801 diraxfile:801 one frame from s01f0008.img.Z. 56 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0 200 peaks in list: Improving=46 Ok=154 Tried=262 Duration=19 Theta=39 Respons=4 peaklist:200 peakfile:955 diraxfile:955 955 reflections written to i1.drx 955 reflections written to i1.pk ! reset display interval graphicsmod 1955 peaks found
view ended at 25-Nov-2010 19:20:58 CPU time used 00:00:11
dirax (version 1.16 2010093000) started on bekken at 25-Nov-2010 19:20:58
Albert J.M. Duisenberg, Indexing in Single-Crystal Diffractometry with an Obstinate List of Reflections
J. Appl. Cryst. (1992). 25,92-96.
C O N D I T I O N S F O R U S E
=====================================
a. You may not copy DirAx neither the auxiliary files except for use by yourself or for use in your laboratory, institute,
office and the like.
b. You may not hand over DirAx in any form to third parties without provable permission from the author.
c. You use DirAx at your own responsibility completely.
Albert J.M. Duisenberg, Antoine M.M. Schreurs
BIJVOET Centre for Biomolecular Research
Laboratory for Crystal and Structural Chemistry
Utrecht University since A.D. MDCXXXVI
Padualaan 8, NL-3584 CH UTRECHT
The Netherlands
Electronic MAIL addresses: a.m.m.schreurs@uu.nl a.j.m.duisenberg@chem.uu.nl
For reproducible results, fix the random generator seed.
Created by view (version 1.4 2010101300) at 25-Nov-2010 19:20:46 Host bekken User schreurs WD /work/schreurs/diamond/diamond/ delta 0.0 -0.11 0.41 detzero 0.0 0.11 -0.41 955 c-vectors from file i1.drx.955 reflections from the input file
145163820 triplets 30000 triplets used Randomizing (Seed=12345) [i,j,k]... 30000 random triplets 29996 triplet vectors Squishd: 29996 t vectors ==> 10930 t vectors Sorting 10930 t vectors... Reducing 10930 t vectors ==> 10841 t vectors Acl #H a b c alpha beta gamma Volume 900 955 57.517 58.154 149.193 89.80 89.50 90.52 498983 371 575 57.507 58.153 145.366 95.96 94.91 90.50 481666 370 578 58.174 57.508 141.862 88.82 95.60 90.54 472216 369 612 58.183 57.512 152.195 88.15 97.71 89.49 504365 349 624 58.171 57.509 152.489 91.95 97.93 90.52 504916 310 522 58.192 57.510 128.374 91.83 100.36 90.54 422351 289 164 48.459 57.507 58.133 90.55 111.83 103.29 145545 252 213 52.848 57.515 58.165 89.46 98.41 96.91 173619 246 359 58.169 57.443 98.389 90.19 106.69 90.67 314889 240 216 57.515 58.192 71.650 109.43 108.53 90.53 212655 142 138 44.962 45.700 57.498 82.03 82.67 79.74 114491 128 166 43.443 57.534 58.110 89.49 104.03 102.72 137303 Solutions limited by voltest selected ACL 900
column 2 (#H) gives the number of fitting reflections
Write the matrix to disk
Dirax>
ccd i1
Sigma calculation stopped after 20000 cells 20000 cell calculations LSQ 57.5168 58.1540 149.1931 89.798 89.503 90.521 498983.313 Mean 57.6512 58.2158 149.3139 89.832 89.620 90.413 497418.281 Max 615.2530 286.2566 899.8871 123.381 169.464 176.951 Min 50.2376 24.5279 138.0338 11.334 42.805 17.943 Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 158.839 Created i1.rmatDirax> exit
dirax ended at 25-Nov-2010 19:21:02 CPU time used 00:00:04
buildsearch started on bekken at 25-Nov-2010 19:21:02 Found Z compressed .img images Available scans: scan s01f 369 images first image=s01f0001.img.Z Total 369 imagesTo refine the cell parameters, and possibly also some experimental parameters, the same 955 reflections from above could be used. But these are all found in frames 1 to 8. It is better to search for more reflections in the whole dataset. (The indexing program will only use 1000 reflections; so that's why the initial search was limited to only a few frames.)
search types: 2: peak search in every frame. The total number of peaks is the number of expected peaks per frame times the number of frames. 3: peak search in first frame. peaksearch in second frame. Select the peaks in the previous frame, if they were stronger. Continue this process for n frames. The total number of peaks will be less than the number of expected peaks per frame times the number of frames. 4: repeat searchtype 3 over many images.search type (2/3/4) [3] 4
Created jsearch.vic s01f From s01f0001.img.Z to s01f0010.img.Z From s01f0041.img.Z to s01f0050.img.Z From s01f0081.img.Z to s01f0090.img.Z From s01f0121.img.Z to s01f0130.img.Z From s01f0161.img.Z to s01f0170.img.Z From s01f0201.img.Z to s01f0210.img.Z From s01f0241.img.Z to s01f0250.img.Z From s01f0281.img.Z to s01f0290.img.Z From s01f0321.img.Z to s01f0330.img.Z buildsearch ended at 25-Nov-2010 19:21:02 CPU time used 00:00:00
viewadscgd (version 1.4 2010101300) started on bekken at 25-Nov-2010 19:21:02Execute the script jsearch.vic, created by buildsearch
! Created by buildsearch at 25-Nov-2010 19:21:02
! Host bekken User schreurs WD /work/schreurs/diamond/diamond/
evpylog searching peaks type 4
! abort on warnings
abort on
! check contiguous images
checknext on
! set number of peaks in one image
peaknr 200
! peak quality
peakbg 3.0
peakbg2 3.0
! forbidden area
durationmax 5.0
! minimum intensity
peakmin 5
! no peaks to be found in next x degrees
peakremove 3.0
! stop after successive weaks
peakbreak 5
! peak size parameters
peaksizemm 0.8
! resolution limits
resomin 40.0
min resolution=40.0 (theta=0.51)
resomax 3.0
max resolution=3.0 (theta=6.8)
! apply new detalign if it exists
&detalign_j
graphics off
! peak file
peakfile j peakopen
Created j.pk
! try read dark image later
trydark on 1.0
! read first image
read s01f0001.img.Z
one frame from s01f0001.img.Z.
Lambda changed from 0.71073 to 1.488 thmin changed to 1.07 thmax changed to 14.36
Initialized DISTOR adsc920 16 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0
Dist Swing Omega Chi Phi Axis Start End
old 0.00 0.00 0.00 0.00 0.00 1
new 116.00 0.00 0.00 0.00 0.00 3 0.00 0.50
! on screen
plot
! initialize peak search
peakstart
! peak search on first image
peak3 sync
BadMaskFill 63181 pixels behind beamstop
Tried=267 Duration=19 Theta=43 Respons=5
peaklist:200 peakfile:0
! peak search on next images
\repeat 9 next plot peak3 sync
one frame from s01f0002.img.Z. 60 overflow pixels. Range 0.0 65535.0 Actual 30.0 5000.0
200 peaks in list: OnFirstFrame=165 Improving=35
Tried=270 Duration=23 Theta=41 Respons=6
....cut....
! close output files
peakclose
11635 reflections written to j.pk
! forget about dark
nodark
! reset display interval
graphicsmod 1
A total of 11635 peaks have been found.
view ended at 25-Nov-2010 19:23:09 CPU time used 00:01:50
peakref (version 1.3 2010112400) started on bekken at 25-Nov-2010 19:23:09read rmatrix, created by dirax
RMAT 1 i1
RMAT DMAT
-0.0045063 -0.0149275 0.0029056 -14.2378531 -43.9840317 34.2180138
-0.0132150 -0.0016800 -0.0042669 -50.2575226 -5.4101052 -28.7540913
0.0103627 -0.0083707 -0.0042757 63.8838196 -96.0092926 -94.6551743
Determinant: 0.2004078E-05 498982.7
Cell: 57.51682 58.15398 149.19292 89.7986 89.5033 90.5206 V= 498982.66
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=-1
rotaxis 0.61151 -0.66932 0.42197 rotang 173.07304
Matrix 1 initial orx and ory shift set to 0.00422
no assumptions here: pointgroup=-1
read the peaks from the second peak search
Peakref>
pk j.pk
FocusType changed to synchrotron ReadPkData: 12181 lines from j.pk 11635 reflections. 72 experiments. n1=0 n2=11635 final residue = +1.0*mmAng +1.0*rotall resfactor 0 1 0 0 0 1Peakref> status
Reind: no reflections changed indices. matrix 1: 11635 reflections.
SetMM 1.0: 416 out of position. SetRot 1.0: 191 out of rot. 11635 reflections (11029 allowed)
Calculating initial residues.
Rmat 1 setting constraints to triclinic.
no constraints for triclinic
Fixing xtalz
One matrix. 11635 reflections. (forbidden mm:416 rot:191 total:606)
Used: 11029 mm 11029 rot
ref current previous change initial change shift
===================================================================
a No 57.51682 57.51682 0.28758
b No 58.15398 58.15398 0.29077
c No 149.19292 149.19292 0.74596
alpha No 89.79857 89.79857 1.00000
beta No 89.50325 89.50325 1.00000
gamma No 90.52058 90.52058 1.00000
orx No 0.61151 0.61151 0.00422
ory No -0.66932 -0.66932 0.00422
ora No 173.07304 173.07304 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes 0.00000 0.00000 0.00000 0.00000 0.00000 0.10000
zerover Yes 0.00000 0.00000 0.00000 0.00000 0.00000 0.10000
detrotx No 0.00000 0.00000 0.20000
detroty No 0.00000 0.00000 0.20000
detrotz No 0.00000 0.00000 0.20000
===================================================================
Vol 498982.59 498982.59 0.00 498982.59 0.00 11029
===================================================================
mm 0.43417 0.43417 0.00000 0.43417 0.00000 11029
mmAng + 0.21445 0.21445 0.00000 0.21445 0.00000 11029
rotoutside 0.27479 0.27479 0.00000 0.27479 0.00000 4372
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 6657
rotall + 0.10890 0.10890 0.00000 0.10890 0.00000 11029
res 0.32335 0.32335 0.00000 0.32335 0.00000
====
Notes
Refining 2 parameters: 9 evaluations in 5 iterations. residue 0.32143 ====
Refining 2 parameters: no evaluations in no iterations. residue 0.32143
Refining 2 parameters: no evaluations in no iterations. residue 0.32143
One matrix. 11635 reflections. (forbidden mm:416 rot:191 total:606)
Used: 11029 mm 11029 rot
ref current previous change initial change shift
=====================================================================
a No 57.51682 57.51682 0.28758
b No 58.15398 58.15398 0.29077
c No 149.19292 149.19292 0.74596
alpha No 89.79857 89.79857 1.00000
beta No 89.50325 89.50325 1.00000
gamma No 90.52058 90.52058 1.00000
orx No 0.61151 0.61151 0.00422
ory No -0.66932 -0.66932 0.00422
ora No 173.07304 173.07304 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes -0.04128 0.00000 -0.04128 0.00000 -0.04128 0.10000
zerover Yes -0.04714 0.00000 -0.04714 0.00000 -0.04714 0.10000
detrotx No 0.00000 0.00000 0.20000
detroty No 0.00000 0.00000 0.20000
detrotz No 0.00000 0.00000 0.20000
=====================================================================
Vol 498982.59 498982.59 0.00 498982.59 0.00 11029
=====================================================================
mm 0.43029 0.43417 -0.00388 0.43417 -0.00388 11029
mmAng + 0.21253 0.21445 -0.00192 0.21445 -0.00192 11029
rotoutside 0.27479 0.27479 0.00000 0.27479 0.00000 4372
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 6657
rotall + 0.10890 0.10890 0.00000 0.10890 0.00000 11029
res 0.32143 0.32335 -0.00192 0.32335 -0.00192
The residue has improved a very little bit. Refine cell parameters and orientation
Refining 11 parameters: 593 evaluations in 410 iterations. residue 0.10011 ====
Refining 11 parameters: 111 evaluations in 64 iterations. residue 0.09983 ====
Refining 11 parameters: 63 evaluations in 37 iterations. residue 0.09981 ====
One matrix. 11635 reflections. (forbidden mm:416 rot:191 total:606)
Used: 11029 mm 11029 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.77696 57.51682 0.26015 57.51682 0.26015 0.28758
b Yes 57.68054 58.15398 -0.47344 58.15398 -0.47344 0.29077
c Yes 149.83981 149.19292 0.64690 149.19292 0.64690 0.74596
alpha Yes 90.04395 89.79857 0.24539 89.79857 0.24539 1.00000
beta Yes 90.04124 89.50325 0.53799 89.50325 0.53799 1.00000
gamma Yes 89.93592 90.52058 -0.58466 90.52058 -0.58466 1.00000
orx Yes 0.60607 0.61151 -0.00544 0.61151 -0.00544 0.00422
ory Yes -0.67159 -0.66932 -0.00226 -0.66932 -0.00226 0.00422
ora Yes 173.08621 173.07304 0.01317 173.07304 0.01317 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes -0.15772 -0.04128 -0.11644 0.00000 -0.15772 0.10000
zerover Yes -0.05039 -0.04714 -0.00326 0.00000 -0.05039 0.10000
detrotx No 0.00000 0.00000 0.20000
detroty No 0.00000 0.00000 0.20000
detrotz No 0.00000 0.00000 0.20000
=====================================================================
Vol 499356.56 498982.59 373.97 498982.59 373.97 11029
=====================================================================
mm 0.14594 0.43064 -0.28470 0.43417 -0.28823 11029
mmAng + 0.07208 0.21270 -0.14062 0.21445 -0.14236 11029
rotoutside 0.20380 0.27535 -0.07155 0.27479 -0.07098 1510
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 9519
rotall + 0.02773 0.10899 -0.08126 0.10890 -0.08117 11029
res 0.09981 0.32169 -0.22188 0.32335 -0.22353
Dramatic improvement. Residue down to 0.10
Refine detector orientation
Peakref>
free detrot
Peakref>
go3
Refining 14 parameters: Amoeba exceeding maximum(500) iterations.
722 evaluations in 500 iterations. (You may want to increase itmax) residue 0.0642 ====
Refining 14 parameters: 313 evaluations in 208 iterations. residue 0.0619 ====
Refining 14 parameters: 119 evaluations in 70 iterations. residue 0.06165 ====
One matrix. 11635 reflections. (forbidden mm:416 rot:191 total:606)
Used: 11029 mm 11029 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.72806 57.77696 -0.04890 57.51682 0.21124 0.28758
b Yes 57.74027 57.68054 0.05973 58.15398 -0.41371 0.29077
c Yes 149.79260 149.83981 -0.04721 149.19292 0.59969 0.74596
alpha Yes 90.01301 90.04395 -0.03095 89.79857 0.21444 1.00000
beta Yes 89.97675 90.04124 -0.06448 89.50325 0.47350 1.00000
gamma Yes 89.97655 89.93592 0.04063 90.52058 -0.54402 1.00000
orx Yes 0.60670 0.60607 0.00064 0.61151 -0.00481 0.00422
ory Yes -0.67136 -0.67159 0.00023 -0.66932 -0.00203 0.00422
ora Yes 173.04492 173.08621 -0.04128 173.07304 -0.02811 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes -0.13695 -0.15772 0.02078 0.00000 -0.13695 0.10000
zerover Yes -0.10134 -0.05039 -0.05095 0.00000 -0.10134 0.10000
detrotx Yes 0.14863 0.00000 0.14863 0.00000 0.14863 0.20000
detroty Yes 0.25508 0.00000 0.25508 0.00000 0.25508 0.20000
detrotz Yes 0.34295 0.00000 0.34295 0.00000 0.34295 0.20000
=====================================================================
Vol 499293.66 499356.56 -62.91 498982.59 311.06 11029
=====================================================================
mm 0.06910 0.14598 -0.07688 0.43417 -0.36507 11029
mmAng + 0.03413 0.07210 -0.03797 0.21445 -0.18032 11029
rotoutside 0.21392 0.20421 0.00971 0.27479 -0.06086 1431
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 9598
rotall + 0.02752 0.02774 -0.00022 0.10890 -0.08138 11029
res 0.06165 0.09984 -0.03819 0.32335 -0.26170
A further improvement. Residue down to 0.06
Refine detector distance
Peakref>
free zerodist
Peakref>
go3
Refining 15 parameters: 258 evaluations in 154 iterations. residue 0.0615 ====
Refining 15 parameters: 186 evaluations in 110 iterations. residue 0.0615 ====
Refining 15 parameters: 126 evaluations in 75 iterations. residue 0.06149 ====
One matrix. 11635 reflections. (forbidden mm:416 rot:191 total:606)
Used: 11029 mm 11029 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.71700 57.72806 -0.01106 57.51682 0.20018 0.28758
b Yes 57.73140 57.74027 -0.00887 58.15398 -0.42258 0.29077
c Yes 149.77315 149.79260 -0.01945 149.19292 0.58023 0.74596
alpha Yes 90.01363 90.01301 0.00063 89.79857 0.21507 1.00000
beta Yes 89.97931 89.97675 0.00255 89.50325 0.47606 1.00000
gamma Yes 89.97445 89.97655 -0.00211 90.52058 -0.54614 1.00000
orx Yes 0.60664 0.60670 -0.00006 0.61151 -0.00487 0.00422
ory Yes -0.67142 -0.67136 -0.00006 -0.66932 -0.00210 0.00422
ora Yes 173.04176 173.04492 -0.00316 173.07304 -0.03127 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist Yes -0.02222 0.00000 -0.02222 0.00000 -0.02222 0.10000
zerohor Yes -0.13514 -0.13695 0.00180 0.00000 -0.13514 0.10000
zerover Yes -0.10342 -0.10134 -0.00208 0.00000 -0.10342 0.10000
detrotx Yes 0.14917 0.14863 0.00053 0.00000 0.14917 0.20000
detroty Yes 0.26663 0.25508 0.01155 0.00000 0.26663 0.20000
detrotz Yes 0.35991 0.34295 0.01695 0.00000 0.35991 0.20000
=====================================================================
Vol 499056.38 499293.66 -237.28 498982.59 73.78 11029
=====================================================================
mm 0.06852 0.06911 -0.00059 0.43417 -0.36565 11029
mmAng + 0.03384 0.03413 -0.00029 0.21445 -0.18061 11029
rotoutside 0.19931 0.21407 -0.01476 0.27479 -0.07548 1543
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 9486
rotall + 0.02765 0.02752 0.00013 0.10890 -0.08125 11029
res 0.06149 0.06166 -0.00016 0.32335 -0.26185
Remarks:
Rmat 1 setting constraints to tetragonal.
Fixing xtalz
final constraints: b=57.7242
SetupRmatrices matrix 1 rotvec= 0.60664 -0.67142 0.42565 AbsVec=1.0 rotang=173.04176
Stored rmat as 1
RMAT 1 i1
RMAT DMAT
-0.0044923 -0.0149537 0.0028924 -14.9687166 -43.8737297 34.3964500
-0.0131671 -0.0016344 -0.0042930 -49.8269386 -5.4460492 -28.6304016
0.0103228 -0.0085923 -0.0042171 64.8812485 -96.2996750 -94.5980606
Determinant: 0.2003781E-05 499056.5
Cell: 57.72419 57.72420 149.77315 90.0000 90.0000 90.0000 V= 499056.53
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/m
a b c alpha beta gamma volume
Refined 1 : 57.724 57.724 149.773 90.00 90.00 90.00 499056.5
Original 1 : 57.517 58.154 149.193 89.80 89.50 90.52 498982.7
Volume ratio = 1.0 No solutions
Pointgroup has been set to 4/m
One matrix. 11635 reflections. (forbidden mm:416 rot:191 total:606)
Used: 11029 mm 11029 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.72420 57.72806 -0.00386 57.51682 0.20738 0.28758
b Con 57.72420 58.15398 0.29077
c Yes 149.77315 149.79260 -0.01945 149.19292 0.58023 0.74596
alpha Fix 90.00000 89.79857 1.00000
beta Fix 90.00000 89.50325 1.00000
gamma Fix 90.00000 90.52058 1.00000
orx Yes 0.60664 0.60670 -0.00006 0.61151 -0.00487 0.00422
ory Yes -0.67142 -0.67136 -0.00006 -0.66932 -0.00210 0.00422
ora Yes 173.04176 173.04492 -0.00316 173.07304 -0.03127 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist Yes -0.02222 0.00000 -0.02222 0.00000 -0.02222 0.10000
zerohor Yes -0.13514 -0.13695 0.00180 0.00000 -0.13514 0.10000
zerover Yes -0.10342 -0.10134 -0.00208 0.00000 -0.10342 0.10000
detrotx Yes 0.14917 0.14863 0.00053 0.00000 0.14917 0.20000
detroty Yes 0.26663 0.25508 0.01155 0.00000 0.26663 0.20000
detrotz Yes 0.35991 0.34295 0.01695 0.00000 0.35991 0.20000
=====================================================================
Vol 499056.53 499293.66 -237.13 498982.59 73.94 11029
=====================================================================
mm 0.07336 0.06911 0.00425 0.43417 -0.36081 11029
mmAng + 0.03623 0.03413 0.00210 0.21445 -0.17822 11029
rotoutside 0.18709 0.21407 -0.02699 0.27479 -0.08770 1692
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 9337
rotall + 0.02854 0.02752 0.00102 0.10890 -0.08035 11029
res 0.06478 0.06166 0.00312 0.32335 -0.25857
Due to the constraints on a, b, alpha, beta and gamma, the
residue increased from 0.061 to 0.065
Refining 11 parameters: 219 evaluations in 139 iterations. residue 0.06267
Refining 11 parameters: 130 evaluations in 77 iterations. residue 0.06256
Refining 11 parameters: 92 evaluations in 54 iterations. residue 0.06251
One matrix. 11635 reflections. (forbidden mm:416 rot:191 total:606)
Used: 11029 mm 11029 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.69807 57.72420 -0.02613 57.51682 0.18125 0.28758
b Con 57.69807 58.15398 0.29077
c Yes 149.70992 149.77315 -0.06323 149.19292 0.51700 0.74596
alpha Fix 90.00000 89.79857 1.00000
beta Fix 90.00000 89.50325 1.00000
gamma Fix 90.00000 90.52058 1.00000
orx Yes 0.60671 0.60664 0.00006 0.61151 -0.00480 0.00422
ory Yes -0.67129 -0.67142 0.00013 -0.66932 -0.00197 0.00422
ora Yes 173.03288 173.04176 -0.00888 173.07304 -0.04015 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist Yes -0.07727 -0.02222 -0.05505 0.00000 -0.07727 0.10000
zerohor Yes -0.13435 -0.13514 0.00079 0.00000 -0.13435 0.10000
zerover Yes -0.10174 -0.10342 0.00167 0.00000 -0.10174 0.10000
detrotx Yes 0.14706 0.14917 -0.00211 0.00000 0.14706 0.20000
detroty Yes 0.26628 0.26663 -0.00035 0.00000 0.26628 0.20000
detrotz Yes 0.35886 0.35991 -0.00105 0.00000 0.35886 0.20000
=====================================================================
Vol 498394.38 499056.53 -662.16 498982.59 -588.22 11029
=====================================================================
mm 0.07161 0.07336 -0.00175 0.43417 -0.36257 11029
mmAng + 0.03537 0.03623 -0.00087 0.21445 -0.17908 11029
rotoutside 0.21148 0.18709 0.02440 0.27479 -0.06330 1425
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 9604
rotall + 0.02714 0.02854 -0.00140 0.10890 -0.08176 11029
res 0.06251 0.06478 -0.00227 0.32335 -0.26084
After all these changes there is a chance that some reflections have been mis-indexed. It is wise to reindex the peaks.
Reind: 813 reflections changed indices. matrix 1: 11635 reflections. SetMM 1.0: 4 out of position. SetRot 1.0: 4 out of rot. 11635 reflections (11630 allowed)813 reflections change indices,
One matrix. 11635 reflections. (forbidden mm:4 rot:4 total:5)
Used: 11630 mm 11630 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.69807 57.72420 -0.02613 57.51682 0.18125 0.28758
b Con 57.69807 58.15398 0.29077
c Yes 149.70992 149.77315 -0.06323 149.19292 0.51700 0.74596
alpha Fix 90.00000 89.79857 1.00000
beta Fix 90.00000 89.50325 1.00000
gamma Fix 90.00000 90.52058 1.00000
orx Yes 0.60671 0.60664 0.00006 0.61151 -0.00480 0.00422
ory Yes -0.67129 -0.67142 0.00013 -0.66932 -0.00197 0.00422
ora Yes 173.03288 173.04176 -0.00888 173.07304 -0.04015 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist Yes -0.07727 -0.02222 -0.05505 0.00000 -0.07727 0.10000
zerohor Yes -0.13435 -0.13514 0.00079 0.00000 -0.13435 0.10000
zerover Yes -0.10174 -0.10342 0.00167 0.00000 -0.10174 0.10000
detrotx Yes 0.14706 0.14917 -0.00211 0.00000 0.14706 0.20000
detroty Yes 0.26628 0.26663 -0.00035 0.00000 0.26628 0.20000
detrotz Yes 0.35886 0.35991 -0.00105 0.00000 0.35886 0.20000
=====================================================================
Vol 498394.38 499056.53 -662.16 498982.59 -588.22 11630
=====================================================================
mm 0.02200 0.07336 -0.05136 0.43417 -0.41217 11630
mmAng + 0.01086 0.03623 -0.02537 0.21445 -0.20358 11630
rotoutside 0.03637 0.18709 -0.15071 0.27479 -0.23841 1022
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 10608
rotall + 0.00317 0.02854 -0.02538 0.10890 -0.10573 11630
res 0.01403 0.06478 -0.05075 0.32335 -0.30932
====
And there are only 5 rejected reflections (two lines above the table).
Refining 11 parameters: 359 evaluations in 232 iterations. residue 0.0128 ====
Refining 11 parameters: 177 evaluations in 106 iterations. residue 0.01277 ====
Refining 11 parameters: 123 evaluations in 73 iterations. residue 0.01276 ====
One matrix. 11635 reflections. (forbidden mm:4 rot:4 total:5)
Used: 11630 mm 11630 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.69087 57.69807 -0.00719 57.51682 0.17406 0.28758
b Con 57.69087 58.15398 0.29077
c Yes 149.68153 149.70992 -0.02838 149.19292 0.48862 0.74596
alpha Fix 90.00000 89.79857 1.00000
beta Fix 90.00000 89.50325 1.00000
gamma Fix 90.00000 90.52058 1.00000
orx Yes 0.60687 0.60671 0.00017 0.61151 -0.00464 0.00422
ory Yes -0.67112 -0.67129 0.00017 -0.66932 -0.00180 0.00422
ora Yes 173.04477 173.03288 0.01188 173.07304 -0.02826 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist Yes -0.09686 -0.07727 -0.01959 0.00000 -0.09686 0.10000
zerohor Yes -0.13563 -0.13435 -0.00128 0.00000 -0.13563 0.10000
zerover Yes -0.10421 -0.10174 -0.00247 0.00000 -0.10421 0.10000
detrotx Yes 0.14892 0.14706 0.00186 0.00000 0.14892 0.20000
detroty Yes 0.27797 0.26628 0.01169 0.00000 0.27797 0.20000
detrotz Yes 0.35661 0.35886 -0.00225 0.00000 0.35661 0.20000
=====================================================================
Vol 498175.53 498394.38 -218.84 498982.59 -807.06 11630
=====================================================================
mm 0.02201 0.02200 0.00001 0.43417 -0.41216 11630
mmAng + 0.01087 0.01086 0.00001 0.21445 -0.20358 11630
rotoutside 0.03250 0.03637 -0.00388 0.27479 -0.24229 685
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 10945
rotall + 0.00189 0.00317 -0.00128 0.10890 -0.10701 11630
res 0.01276 0.01403 -0.00127 0.32335 -0.31059
And keep the results
Adding original position(0.0 0.11 -0.41) to refined detzero.Enter name [detalign.vic] detalign.vic
WARNING: renaming existing file detalign.vic into detalign.vic.~2~ Created detalign.vicEnter name [goniostat.vic] goniostat.vic
Created goniostat.vic
initial new
-0.0045063 -0.0149275 0.0029056 -0.0044853 -0.0149614 0.0028971
-0.0132150 -0.0016800 -0.0042669 -0.0131739 -0.0016492 -0.0042951
0.0103627 -0.0083707 -0.0042757 0.0103339 -0.0085963 -0.0042181
Determinant: 0.2004078E-05 0.2007325E-05
SameRmat=false
Enter name [i1r]
ic.rmat
Created ic.rmatRemember, the pointgroup has been set to 4/m. The file ic.rmat contains this information.
peakref ended at 25-Nov-2010 19:23:36 CPU time used 00:00:27
builddatcol started on bekken at 25-Nov-2010 19:23:36script name [datcol] datcol
datcoltemplate=datcol Found Z compressed .img imagesload the cell matrix, saved by peakref
rmat ic.rmat subdir ic Created datcolsetup.vicMinimum resolution (angstrom)[50.0] 50
searching *.img* new scan:s01f 369 images Created datcol.vic Created datcols01f.vic builddatcol ended at 25-Nov-2010 19:23:36 CPU time used 00:00:00
viewadscgd (version 1.4 2010101300) started on bekken at 25-Nov-2010 19:23:36 Goniostat: type horax id adsc920create the boxfiles by executing the previously build script.
! Created by builddatcol at 25-Nov-2010 19:23:36
! Host bekken User schreurs WD /work/schreurs/diamond/diamond/
evpylog creating boxfiles
@datcols01f.vic
one frame from s01f0001.img.Z.
Lambda changed from 0.71073 to 1.488
Initialized DISTOR adsc920 No files badpixel.* found in [.:adsc920:/usr/global/calibration/adsc920] 16 overflow pixels.
Filling scratch for CorrectBadPixels. 0 same 0 replaced. Range 0.0 65535.0 Actual 30.0 5000.0
Dist Swing Omega Chi Phi Axis Start End
old 0.00 0.00 0.00 0.00 0.00 1
new 116.00 0.00 0.00 0.00 0.00 3 0.00 0.50
RMAT 1 ic
RMAT DMAT
-0.0044853 -0.0149614 0.0028971 -14.9281244 -43.8460999 34.3934746
-0.0131739 -0.0016492 -0.0042951 -49.7948074 -5.4893670 -28.6107864
0.0103339 -0.0085963 -0.0042181 64.9072723 -96.2312393 -94.5056610
Determinant: 0.2007317E-05 498177.4
Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/m
min resolution=50.0 (theta=0.85)
max resolution=1.6 (theta=27.71)
boxsizemm=2.3 boxhor=23 boxver=23 boxang=1.14
Setting ZoomWindow 382 382
DatcolSetup Resolution 50.0 to 1.6 (Theta 0.85 to 27.71). Rotrange from -1.0 to 359.0
rmat h k l genrtd presnt theta dura left rotran impact right rotran impact total
1 36 36 93 996523 996522 474834 463788 463788 463788 453603 463788 463788 454900 908503
Behind Beamstop:10 Total accepted:908493
too many: 808493 reflections (out of 908493). 11.01% useable. scanlength reduced from 360.0 to 33.16
Activescanlength reduced from 360.0 to 35.66 datcolrange1 -1.0 datcolrange2 changed to 34.66
DatcolSetup Resolution 50.0 to 1.6 (Theta 0.85 to 27.71). Rotrange from -1.0 to 34.66
rmat h k l genrtd presnt theta dura left rotran impact right rotran impact total
1 36 36 93 996523 996522 474834 463788 463788 45966 44941 463788 45931 45095 90036
Behind Beamstop:1 Total accepted:90035
Created ic/s01f001.shoe (3817) + 0 = 0

s01f0002 (5064) + 0 = 0 s01f0003 (5040) + 1264 = 1264 Closed ic/s01f001.shoe compress <ic/s01f001.shoe >ic/s01f001.shoe.Z && rm ic/s01f001.shoe s01f0004 Created ic/s01f002.shoe (5043) + 1255 = 1255 Closed ic/s01f002.shoe compress <ic/s01f002.shoe >ic/s01f002.shoe.Z && rm ic/s01f002.shoe s01f0005 Created ic/s01f003.shoe (5029) + 1298 = 1298 Closed ic/s01f003.shoe compress <ic/s01f003.shoe >ic/s01f003.shoe.Z && rm ic/s01f003.shoe s01f0006 Created ic/s01f004.shoe (5028) + 1247 = 1247 Closed ic/s01f004.shoe compress <ic/s01f004.shoe >ic/s01f004.shoe.Z && rm ic/s01f004.shoe s01f0007 Created ic/s01f005.shoe (5029) + 1239 = 1239 Closed ic/s01f005.shoe compress <ic/s01f005.shoe >ic/s01f005.shoe.Z && rm ic/s01f005.shoe s01f0008 Created ic/s01f006.shoe (5067) + 1258 = 1258 Closed ic/s01f006.shoe compress <ic/s01f006.shoe >ic/s01f006.shoe.Z && rm ic/s01f006.shoe s01f0009 Created ic/s01f007.shoe (5021) + 1283 = 1283 Closed ic/s01f007.shoe compress <ic/s01f007.shoe >ic/s01f007.shoe.Z && rm ic/s01f007.shoe s01f0010 Created ic/s01f008.shoe (4969) + 1245 = 1245 Closed ic/s01f008.shoe compress <ic/s01f008.shoe >ic/s01f008.shoe.Z && rm ic/s01f008.shoe s01f0011 Created ic/s01f009.shoe (5013) + 1241 = 1241 Closed ic/s01f009.shoe ....cut.... compress <ic/s01f363.shoe >ic/s01f363.shoe.Z && rm ic/s01f363.shoe s01f0366 Created ic/s01f364.shoe (5028) + 1248 = 1248 Closed ic/s01f364.shoe compress <ic/s01f364.shoe >ic/s01f364.shoe.Z && rm ic/s01f364.shoe s01f0367 Created ic/s01f365.shoe (5028) + 1240 = 1240 Closed ic/s01f365.shoe compress <ic/s01f365.shoe >ic/s01f365.shoe.Z && rm ic/s01f365.shoe s01f0368 Created ic/s01f366.shoe (5067) + 1258 = 1258 Closed ic/s01f366.shoe compress <ic/s01f366.shoe >ic/s01f366.shoe.Z && rm ic/s01f366.shoe s01f0369 Created ic/s01f367.shoe (5023) + 1283 = 1283 Closed ic/s01f367.shoe compress <ic/s01f367.shoe >ic/s01f367.shoe.Z && rm ic/s01f367.shoe Created ic/s01f368.shoe (0) + 2486 = 2486 Closed ic/s01f368.shoe compress <ic/s01f368.shoe >ic/s01f368.shoe.Z && rm ic/s01f368.shoe view ended at 25-Nov-2010 19:43:40 CPU time used 00:10:37
buildeval15 (version 1.1 2010112500) started on bekken at 25-Nov-2010 19:43:41 spectrum s01f001.shoe.Z Opened learning: detectortype=adsc imagedepth=0.5 lambda=1.488 detid=adsc920 gonid=adsc920 calid=adsc920 s01f001.shoe.Z closed after 8192 bytes 47 lines Created spectrum.pic lambda 1.488 focus no xtal.evc found. Created focus.pic Possible Focus Types: unknown tube rotating mirror synchrotron fileInitiate some variables. The information needed will be extracted from the first boxfile or possibly available other files. Missing information has to be supplied by the user.
detector Created detector.picpointspreadgamma[0.8] 0.8
crystal no p4p files found. no absorb.ins found. no xtal.evc found. Created crystal.picWe don't known anything about the crystal. Let's assume it is a small sphere with 0.1 mm diameter.
mosaic no xtal.evc found. Created mosaic.picWe don't known anything about the mosaicity. Inspection of the images we have seen until now suggests a small value.
simulation Created simulation.pic refine Created refine.pic refineshift exrot => 0.1 Created eval15.init buildeval15 ended at 25-Nov-2010 19:43:41 CPU time used 00:00:00ic> cd -
eval15 (version 1.2 2010112500) started on bekken at 25-Nov-2010 19:43:41 lambdaadd 1 1.48800 0.00010 1.00 lambdacalcmean: 1.488(0.0001,1.0) Mean=1.488 Mid=1.488 SetupXtalModel 4: 20 corners 12 new faces ( 1 2 3 4 5 6 7 8 9 10 11 12 ).The reflections in the first box file originate from the first image. Part of these reflections would probably have impacted the non-existing previous frame. Therefore the first reflections in the first boxfile will probably be incomplete. So open the third boxfile, and read reflection number 1.
s01f003.shoe.Z Opened ignoring focus parameters ALPHA 1.488 (ignored) Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41 No FaceRmat 12 faces 20 corners 30 edgesThe wavelength has been set by the file spectrum.pic. The value contained in the boxfile is ignored.
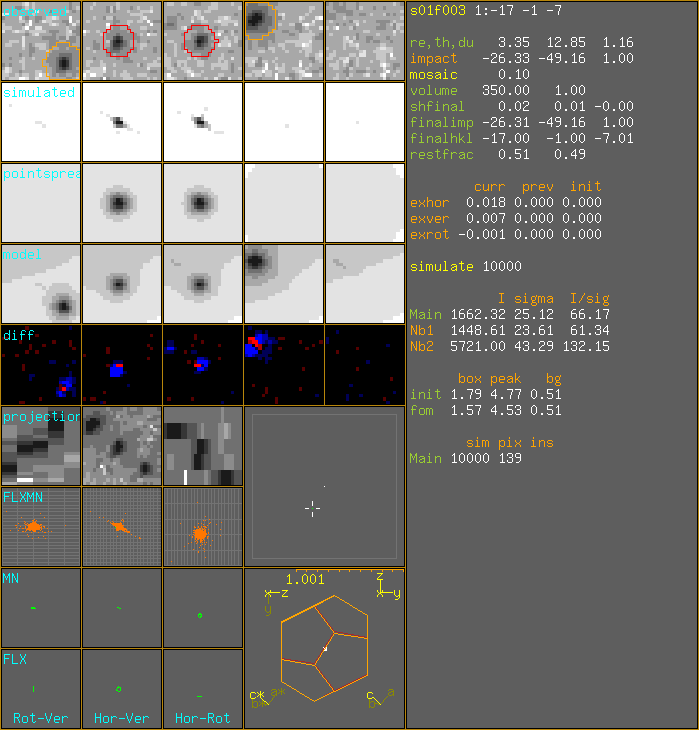
SampleXtal SampleSize 0.01 mm. xtaldim 0.093 0.098 0.079 350 sample points. volume = 350.0 mu^3 1:-17 -1 -7 sim:10000m px:138m DrawSimulatedImpacts I=1606.048 s=24.257 I/s=66.21 nb h k l hor ver rot dhor dver drot np Intensity Sigma I/sig SI SS SI/SS 1 -17 -1 -8 -25.54 -49.85 0.24 0.79 -0.69 -0.76 9974 1433.52 23.10 62.1 2 -17 -1 -6 -27.11 -48.49 1.77 -0.79 0.67 0.77 9994 5696.58 41.70 136.6 I=1606.048 s=24.257 I/s=66.21 foms: 1.791 4.774 0.51
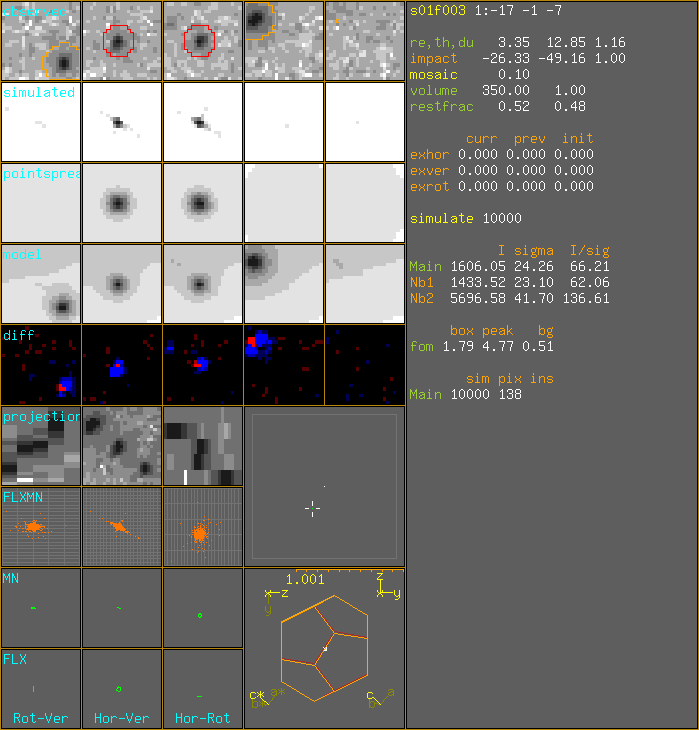
1:-17 -1 -7 DrawSimulatedImpacts I=1605.995 s=24.251 I/s=66.223 nb h k l hor ver rot dhor dver drot np Intensity Sigma I/sig SI SS SI/SS 1 -17 -1 -8 -25.54 -49.85 0.24 0.79 -0.69 -0.76 9974 1433.80 23.10 62.1 2 -17 -1 -6 -27.11 -48.49 1.77 -0.79 0.67 0.77 9994 5695.99 41.69 136.6 I=1605.995 s=24.251 I/s=66.223 foms: 1.791 4.774 0.51 30/60 RefFom=1.566 1:-17 -1 -7 sim:10000m px:139m DrawSimulatedImpacts I=1662.317 s=25.121 I/s=66.173 nb h k l hor ver rot dhor dver drot np Intensity Sigma I/sig SI SS SI/SS 1 -17 -1 -8 -25.54 -49.85 0.24 0.79 -0.69 -0.76 9974 1448.61 23.61 61.3 2 -17 -1 -6 -27.11 -48.49 1.77 -0.79 0.67 0.77 9994 5721.00 43.29 132.2 I=1662.317 s=25.121 I/s=66.173 foms: 1.565 4.533 0.505 FinalImpact -26.308 -49.155 1.0 variable value last init shift ========================================= exhor 0.01778 0.00000 0.00000 0.11000 exver 0.00697 0.00000 0.00000 0.11000 exrot -0.00142 0.00000 0.00000 0.10000 ========================================= box 1.56510 1.79116 1.79116 peak 4.53314 4.77422 4.77422 bg 0.50535 0.50964 0.50964 FinalImpact -26.308 -49.155 1.0
Try another sigma for the wavelength
Refl nr [2]
lambdasigma
Let's try a ten times larger value
lambdasigma1[0.0001]
*10
Refl nr [2]
go
1:-17 -1 -7 sim:9993m+7e px:154m DrawSimulatedImpacts I=1987.554 s=29.203 I/s=68.06 nb h k l hor ver rot dhor dver drot np Intensity Sigma I/sig SI SS SI/SS 1 -17 -1 -8 -25.54 -49.85 0.24 0.79 -0.69 -0.76 9946 1574.02 25.55 61.6 2 -17 -1 -6 -27.11 -48.49 1.77 -0.79 0.67 0.77 9956 5946.91 45.09 131.9 I=1987.554 s=29.203 I/s=68.06 foms: 1.012 2.431 0.487 FinalImpact -26.308 -49.155 1.0
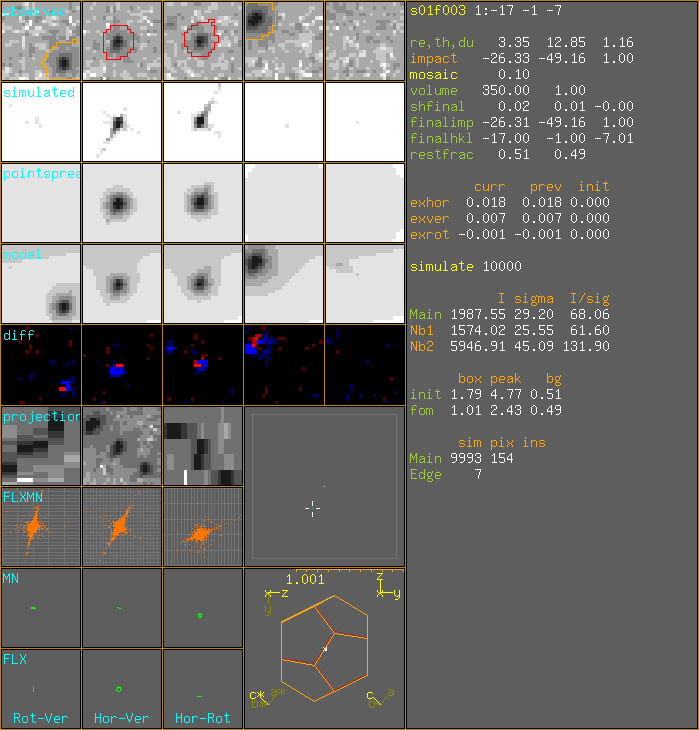
WARNING: renaming existing file spectrum.pic into spectrum.pic.~1~ Created spectrum.picRefl nr [2] exit
64 go_runs. eval15 ended at 25-Nov-2010 19:43:42 CPU time used 00:00:01ic> cd -
build todo list
ic>
batchsetup set
independent run for each boxfile. 368 jobs todo building computers... name avail load n ecm6 bekken 2.13 2.87 4 2 cello 0.71 1.19 1 1 cornet 4.00 2.08 4 4 gong 3.01 3.99 4 4 harp 1.00 0.80 1 1 klok 3.17 3.83 4 4 tamtam 3.66 2.34 4 3 triangel -9.02 12.02 4 0 trommel -5.00 8.00 4 0 vibrafoon -1.11 4.11 4 0 xylofoon 3.16 3.84 4 4 11 hosts 38 cpu's. Total load = 45.07 21 useable cpu's cornet tamtam klok xylofoon gong cornet tamtam klok xylofoon bekken gong cornet tamtam klok xylofoon bekken gong cornet harp cello tamtam You may want to change the file computers. Start the batch proces with batchstart.ic> batchstart
21 CPU's to be used
number of jobs = 368
number of simultaneous jobs = 21
available number of rows = 8
number of columns = 3
...(cut)....
21:31:24 done:367 busy:1
21:31:44
number of jobs done = 368
Created final.y
s01f s01f001.y s01f002.y s01f003.y s01f004.y s01f005.y s01f006.y s01f007.y
s01f008.y s01f009.y s01f010.y s01f011.y s01f012.y s01f013.y s01f014.y
s01f015.y s01f016.y s01f017.y s01f018.y s01f019.y s01f020.y s01f021.y
s01f022.y s01f023.y s01f024.y s01f025.y s01f026.y s01f027.y s01f028.y
s01f029.y s01f030.y s01f031.y s01f032.y s01f033.y s01f034.y s01f035.y
s01f036.y s01f037.y s01f038.y s01f039.y s01f040.y s01f041.y s01f042.y
s01f043.y s01f044.y s01f045.y s01f046.y s01f047.y s01f048.y s01f049.y
s01f050.y s01f051.y s01f052.y s01f053.y s01f054.y s01f055.y s01f056.y
s01f057.y s01f058.y s01f059.y s01f060.y s01f061.y s01f062.y s01f063.y
s01f064.y s01f065.y s01f066.y s01f067.y s01f068.y s01f069.y s01f070.y
s01f071.y s01f072.y s01f073.y s01f074.y s01f075.y s01f076.y s01f077.y
s01f078.y s01f079.y s01f080.y s01f081.y s01f082.y s01f083.y s01f084.y
s01f085.y s01f086.y s01f087.y s01f088.y s01f089.y s01f090.y s01f091.y
s01f092.y s01f093.y s01f094.y s01f095.y s01f096.y s01f097.y s01f098.y
s01f099.y s01f100.y s01f101.y s01f102.y s01f103.y s01f104.y s01f105.y
s01f106.y s01f107.y s01f108.y s01f109.y s01f110.y s01f111.y s01f112.y
s01f113.y s01f114.y s01f115.y s01f116.y s01f117.y s01f118.y s01f119.y
s01f120.y s01f121.y s01f122.y s01f123.y s01f124.y s01f125.y s01f126.y
s01f127.y s01f128.y s01f129.y s01f130.y s01f131.y s01f132.y s01f133.y
s01f134.y s01f135.y s01f136.y s01f137.y s01f138.y s01f139.y s01f140.y
s01f141.y s01f142.y s01f143.y s01f144.y s01f145.y s01f146.y s01f147.y
s01f148.y s01f149.y s01f150.y s01f151.y s01f152.y s01f153.y s01f154.y
s01f155.y s01f156.y s01f157.y s01f158.y s01f159.y s01f160.y s01f161.y
s01f162.y s01f163.y s01f164.y s01f165.y s01f166.y s01f167.y s01f168.y
s01f169.y s01f170.y s01f171.y s01f172.y s01f173.y s01f174.y s01f175.y
s01f176.y s01f177.y s01f178.y s01f179.y s01f180.y s01f181.y s01f182.y
s01f183.y s01f184.y s01f185.y s01f186.y s01f187.y s01f188.y s01f189.y
s01f190.y s01f191.y s01f192.y s01f193.y s01f194.y s01f195.y s01f196.y
s01f197.y s01f198.y s01f199.y s01f200.y s01f201.y s01f202.y s01f203.y
s01f204.y s01f205.y s01f206.y s01f207.y s01f208.y s01f209.y s01f210.y
s01f211.y s01f212.y s01f213.y s01f214.y s01f215.y s01f216.y s01f217.y
s01f218.y s01f219.y s01f220.y s01f221.y s01f222.y s01f223.y s01f224.y
s01f225.y s01f226.y s01f227.y s01f228.y s01f229.y s01f230.y s01f231.y
s01f232.y s01f233.y s01f234.y s01f235.y s01f236.y s01f237.y s01f238.y
s01f239.y s01f240.y s01f241.y s01f242.y s01f243.y s01f244.y s01f245.y
s01f246.y s01f247.y s01f248.y s01f249.y s01f250.y s01f251.y s01f252.y
s01f253.y s01f254.y s01f255.y s01f256.y s01f257.y s01f258.y s01f259.y
s01f260.y s01f261.y s01f262.y s01f263.y s01f264.y s01f265.y s01f266.y
s01f267.y s01f268.y s01f269.y s01f270.y s01f271.y s01f272.y s01f273.y
s01f274.y s01f275.y s01f276.y s01f277.y s01f278.y s01f279.y s01f280.y
s01f281.y s01f282.y s01f283.y s01f284.y s01f285.y s01f286.y s01f287.y
s01f288.y s01f289.y s01f290.y s01f291.y s01f292.y s01f293.y s01f294.y
s01f295.y s01f296.y s01f297.y s01f298.y s01f299.y s01f300.y s01f301.y
s01f302.y s01f303.y s01f304.y s01f305.y s01f306.y s01f307.y s01f308.y
s01f309.y s01f310.y s01f311.y s01f312.y s01f313.y s01f314.y s01f315.y
s01f316.y s01f317.y s01f318.y s01f319.y s01f320.y s01f321.y s01f322.y
s01f323.y s01f324.y s01f325.y s01f326.y s01f327.y s01f328.y s01f329.y
s01f330.y s01f331.y s01f332.y s01f333.y s01f334.y s01f335.y s01f336.y
s01f337.y s01f338.y s01f339.y s01f340.y s01f341.y s01f342.y s01f343.y
s01f344.y s01f345.y s01f346.y s01f347.y s01f348.y s01f349.y s01f350.y
s01f351.y s01f352.y s01f353.y s01f354.y s01f355.y s01f356.y s01f357.y
s01f358.y s01f359.y s01f360.y s01f361.y s01f362.y s01f363.y s01f364.y
s01f365.y s01f366.y s01f367.y s01f368.y
368 files. 4719553 lines input. 15781 lines skipped. 4703405 lines output.
ic>
cd -
any (version 1.5 2010111000) started on bekken at 25-Nov-2010 21:32:29 Created any.logAny> read final
y-data from final.y
465397 reflections nPg=8 (h,k,l) (k,-h,l) (-h,-k,l) (-k,h,l) (-h,-k,-l) (-k,h,-l) (h,k,-l) (k,-h,-l)
Start mdian2 on 465397 reflections mdian2 stopped after 101 passes median=31956.0 presentationscale=0.0000626
Calculating Extremes
Cannot calculate icr for reflection 54. (total 1853 icr's failed)
69 too negative reflections (allowed) 34626 weak reflections (allowed)
Status for final.y
RMAT 1
RMAT DMAT
-0.0044853 -0.0149614 0.0028971 -14.9281244 -43.8460999 34.3934746
-0.0131739 -0.0016492 -0.0042951 -49.7948074 -5.4893670 -28.6107864
0.0103339 -0.0085963 -0.0042181 64.9072723 -96.2312393 -94.5056610
Determinant: 0.2007317E-05 498177.4
Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/m
Lambda=1.488 pointgroup=4/m
reflections=465397 unique=64247 weak=34626 negative=69
Unique: H from -36 to 0 K from -25 to 24 L from -93 to 0
Full Sphere: H from -36 to 36 K from -36 to 36 L from -93 to 93
Max equivalents 11 datalimit theta 0.8544 27.7102. datalimit reso 1.6 49.894. datalimit chi 0.0 78.1585.
datalimit psi -153.2682 153.0809.
datalimit intensity -108.0624 16449.5723. datalimit duration 1.0 5.0.
one Experiment 368 Sets 369 Images
presentationscale 0.0000626
Selectable: GOOD WEAK EDGEVER EDGEHOR EDGEROT BIGHOR NEGATIVE BADUNIF OVERFLOW
Forbid: EDGEVER EDGEHOR EDGEROT BIGHOR BADUNIF OVERFLOW
Allow: GOOD WEAK NEGATIVE
Require: NONE
Rsym=0.097 Rmeas=0.105 Rpim=0.039 Chi2=28.511 nRsym=453087 Unique1=45 Unique2+=64113
<I>=194.609 <s>=4.871 <I/s>=27.387 <I>/<s>=39.95 nMean=453132
The overal rsym (see the last 2 lines) is not that good.
But note, these data are unscaled.
Print rsym and completeness for various resolution shells.
Any>
rshell
-------------------------------------------------------------------------------- Completeness and Rmerge for Shells Forbid: EDGEVER EDGEHOR EDGEROT BIGHOR BADUNIF OVERFLOW Allow: GOOD WEAK NEGATIVE Require: NONE Sh Theta Reso Meas Equi Obs Mis Total Perc Cum Uni1 Uni2+ Nrsym Redun Rsym Rmeas Rpim Chi2 1 12.46 3.45 43890 6554 50444 638 51082 98.8 98.8 40 6396 47637 7.45 0.068 0.074 0.028 26.61 2 15.78 2.74 41908 8818 50726 0 50726 100.0 99.4 2 6411 47178 7.36 0.090 0.097 0.036 38.66 3 18.14 2.39 41283 9627 50910 2 50912 100.0 99.6 0 6424 47538 7.40 0.119 0.128 0.047 46.48 4 20.04 2.17 40534 10578 51112 0 51112 100.0 99.7 0 6442 47429 7.36 0.111 0.119 0.044 34.46 5 21.66 2.02 39514 11474 50988 0 50988 100.0 99.7 0 6422 46840 7.29 0.116 0.125 0.046 29.16 6 23.09 1.90 38439 12269 50708 0 50708 100.0 99.8 0 6382 46124 7.23 0.130 0.140 0.052 25.40 7 24.39 1.80 37947 12947 50894 0 50894 100.0 99.8 0 6405 45920 7.17 0.145 0.157 0.059 19.75 8 25.57 1.72 37525 13769 51294 0 51294 100.0 99.8 0 6453 45886 7.11 0.165 0.178 0.067 21.19 9 26.68 1.66 36422 14280 50702 0 50702 100.0 99.9 0 6376 44911 7.04 0.183 0.198 0.075 22.50 10 27.71 1.60 26931 23987 50918 20 50938 100.0 99.9 3 6401 33622 5.25 0.203 0.227 0.098 16.53 ======================================================================================================== 27.71 1.60 384393 124303 508696 660 509356 99.9 99.9 45 64112 453085 7.07 0.097 0.105 0.039 28.51The geometry allows both 4/mmm and 4/m. Is this reflected by the symmetry of the intensities?
pg R Rmeas Rpim Chi2 Uni1 Uni2+ nRsym -1 0.078 0.109 0.076 30.07 5877 218373 447255 -1 Triclinic 2/m 0.092 0.109 0.058 30.04 918 127386 452214 2/m Monoclinic mmm 0.096 0.104 0.039 27.74 79 66632 453053 mmm Orthorhombic 4/m 0.097 0.105 0.039 28.51 45 64113 453087 4/m Tetragonal low 4/mmm 0.098 0.102 0.028 27.47 27 34315 453105 4/mmm Tetragonal high -3 0.579 0.648 0.287 2406.36 904 123017 452228 -3 Trigonal low -3m1 0.585 0.619 0.198 2210.61 114 65028 453018 -3m1 Trigonal high -31m 0.595 0.629 0.202 2333.14 117 65031 453015 -31m Trigonal high 6/m 0.596 0.628 0.199 2318.83 93 63741 453039 6/m Hexagonal low 6/mmm 0.598 0.616 0.143 2230.06 14 34267 453118 6/mmm Hexagonal high R-3 0.523 0.584 0.254 2289.60 2597 146934 450535 R-3 Rhombohedral low R-3m 0.530 0.560 0.178 2083.88 478 85543 452654 R-3m Rhombohedral high m3 0.532 0.562 0.176 2104.33 2524 83034 450608 m3 Cubic low m3m 0.535 0.543 0.090 1948.21 4 23491 453128 m3m Cubic high Building 4/mYes. So set pointgroup to 4/mmm.
pointgroup set to 4/mmm Calculating Extremes Cannot calculate icr for reflection 54. (total 1372 icr's failed) nPg=16 (h,k,l) (k,-h,l) (-h,-k,l) (-k,h,l) (h,-k,-l) (-k,-h,-l) (-h,k,-l) (k,h,-l) (-h,-k,-l) (-k,h,-l) (h,k,-l) (k,-h,-l) (-h,k,l) (k,h,l) (h,-k,l) (-k,-h,l)Any> rshell
-------------------------------------------------------------------------------- Completeness and Rmerge for Shells Forbid: EDGEVER EDGEHOR EDGEROT BIGHOR BADUNIF OVERFLOW Allow: GOOD WEAK NEGATIVE Require: NONE Sh Theta Reso Meas Equi Obs Mis Total Perc Cum Uni1 Uni2+ Nrsym Redun Rsym Rmeas Rpim Chi2 1 12.46 3.45 43890 6634 50524 558 51082 98.9 98.9 25 3670 47652 12.98 0.069 0.072 0.020 26.23 2 15.78 2.74 41908 8818 50726 0 50726 100.0 99.5 2 3491 47178 13.51 0.091 0.094 0.025 37.48 3 18.14 2.39 41283 9627 50910 2 50912 100.0 99.6 0 3454 47538 13.76 0.119 0.124 0.033 44.75 4 20.04 2.17 40534 10578 51112 0 51112 100.0 99.7 0 3436 47429 13.80 0.112 0.116 0.031 32.64 5 21.66 2.02 39514 11474 50988 0 50988 100.0 99.8 0 3411 46840 13.73 0.117 0.122 0.033 28.14 6 23.09 1.90 38439 12269 50708 0 50708 100.0 99.8 0 3374 46124 13.67 0.131 0.136 0.037 24.37 7 24.39 1.80 37947 12947 50894 0 50894 100.0 99.8 0 3377 45920 13.60 0.147 0.152 0.041 19.05 8 25.57 1.72 37525 13769 51294 0 51294 100.0 99.9 0 3394 45886 13.52 0.167 0.174 0.047 20.35 9 26.68 1.66 36422 14280 50702 0 50702 100.0 99.9 0 3347 44911 13.42 0.188 0.195 0.053 21.53 10 27.71 1.60 26931 24007 50938 0 50938 100.0 99.9 0 3360 33625 10.01 0.211 0.223 0.070 15.88 ======================================================================================================== 27.71 1.60 384393 124403 508796 560 509356 99.9 99.9 27 34314 453103 13.20 0.098 0.102 0.028 27.47
Print the ratio between the intensity of each reflection divided by
the average intensity of the group of corresponding equivalent reflections.
Any>
icrscale 5
Any>
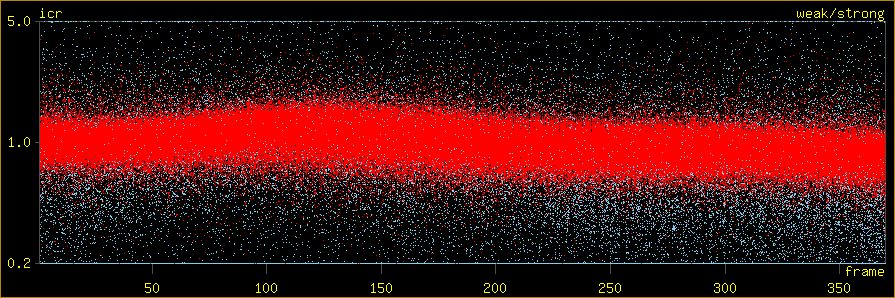
icr
LoadColour horizontal: datalimit frame 0.837 370.208. plotlimit frame 0.837 370.208.
horizontal: datalimit frame 0.837 370.208. plotlimit frame 0.837 370.208. vertical: datalimit shifthor -0.756 0.605. plotlimit shifthor -0.756 0.605. 420395 points
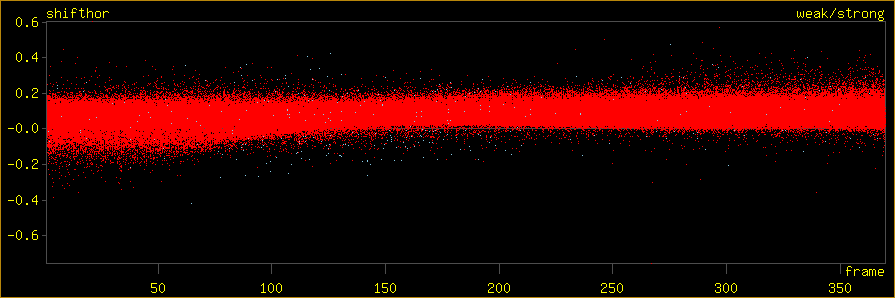
Write the final impacts to a file. The difference between observed and
expected position can be used for a further refinement.
pkrestfrac will be used to filter out all reflections which lie mostly
within one frame.
Any>
pkrestfrac 0.2
Any>
pk
Created final.pk allow=453132 shifted=420395 2dim=328547 3dim=91848. 420395 lines written. Closed final.pkAny> exit
any ended at 25-Nov-2010 21:33:41 CPU time used 00:01:10ic> cd -
peakref (version 1.3 2010112400) started on bekken at 25-Nov-2010 21:33:41Do a postrefinement and calculate sigma's in the cell parameters
Read the orientation matrix.
Peakref>
rmat ic
RMAT 1 ic
RMAT DMAT
-0.0044853 -0.0149614 0.0028971 -14.9281244 -43.8460999 34.3934746
-0.0131739 -0.0016492 -0.0042951 -49.7948074 -5.4893670 -28.6107864
0.0103339 -0.0085963 -0.0042181 64.9072723 -96.2312393 -94.5056610
Determinant: 0.2007317E-05 498177.4
Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/m
rotaxis 0.60687 -0.67112 0.42579 rotang 173.04532
Matrix 1 initial orx and ory shift set to 0.00426
The pointgroup is still set to 4/m. We don't have to change this
into 4/mmm because the constraints will be the same.
Load the refined reflection positions from eval15.
Peakref>
pk ic/final
FocusType changed to synchrotron 91848 hkl records 328547 hkl2 records ReadPkData: 423722 lines from ic/final.pk 420395 reflections. one experiment. detzero: 368 equal records. detrot: 368 equal records. n1=91848 n2=328547 final residue = +1.0*mmAng +1.0*rotpartial resfactor 0 1 1 0 0 0Peakref> status
Reind: No indexing of hkl data.
SetMM 1.0: one out of position. SetWeak 0.0: 43 weak. 420395 reflections (420351 allowed)
Calculating initial residues.
Rmat 1 setting constraints to tetragonal.
Fixing xtalz
One matrix. 420395 reflections. (forbidden mm:1 weak:43 total:44)
Used: 420351 mm 91845 rot
ref current previous change initial change shift
=====================================================================
a No 57.69087 57.69090 0.28845
b Con 57.69087 57.69084 0.28845
c No 149.68208 149.68208 0.74841
alpha Fix 90.00000 89.99943 1.00000
beta Fix 90.00000 89.99971 1.00000
gamma Fix 90.00000 89.99989 1.00000
orx No 0.60687 0.60687 0.00426
ory No -0.67112 -0.67112 0.00426
ora No 173.04532 173.04532 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes 0.00000 0.00000 0.00000 0.00000 0.00000 0.10000
zerover Yes 0.00000 0.00000 0.00000 0.00000 0.00000 0.10000
detrotx No 0.00000 0.00000 0.20000
detroty No 0.00000 0.00000 0.20000
detrotz No 0.00000 0.00000 0.20000
=====================================================================
Vol 498177.31 498177.34 -0.03 498177.34 -0.03 420351
=====================================================================
mm 0.11102 0.11092 0.00009 0.11092 0.00009 420351
mmAng + 0.05488 0.05483 0.00005 0.05483 0.00005 420351
rotpartial + 0.04904 0.04906 -0.00002 0.04906 -0.00002 91845
rotoutside 0.12553 0.12527 0.00026 0.12527 0.00026 6
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 328500
rotall 0.01072 0.01072 -0.00000 0.01072 -0.00000 420351
res 0.10392 0.10389 0.00003 0.10389 0.00003
====
Refine the detector position
Peakref>
go3
Refining 2 parameters: 24 evaluations in 12 iterations. residue 0.08024 ====
Refining 2 parameters: 12 evaluations in 6 iterations. residue 0.08025
Refining 2 parameters: no evaluations in no iterations. residue 0.08025
One matrix. 420395 reflections. (forbidden mm:1 weak:43 total:44)
Used: 420351 mm 91845 rot
ref current previous change initial change shift
=====================================================================
a No 57.69087 57.69090 0.28845
b Con 57.69087 57.69084 0.28845
c No 149.68208 149.68208 0.74841
alpha Fix 90.00000 89.99943 1.00000
beta Fix 90.00000 89.99971 1.00000
gamma Fix 90.00000 89.99989 1.00000
orx No 0.60687 0.60687 0.00426
ory No -0.67112 -0.67112 0.00426
ora No 173.04532 173.04532 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes 0.06692 0.00000 0.06692 0.00000 0.06692 0.10000
zerover Yes 0.05692 0.00000 0.05692 0.00000 0.05692 0.10000
detrotx No 0.00000 0.00000 0.20000
detroty No 0.00000 0.00000 0.20000
detrotz No 0.00000 0.00000 0.20000
=====================================================================
Vol 498177.31 498177.31 0.00 498177.34 -0.03 420351
=====================================================================
mm 0.06313 0.11102 -0.04788 0.11092 -0.04779 420351
mmAng + 0.03121 0.05488 -0.02367 0.05483 -0.02363 420351
rotpartial + 0.04904 0.04904 0.00000 0.04906 -0.00002 91845
rotoutside 0.12553 0.12553 0.00000 0.12527 0.00026 6
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 328500
rotall 0.01072 0.01072 0.00000 0.01072 -0.00000 420351
res 0.08025 0.10392 -0.02367 0.10389 -0.02364
Refine the cell parameters and orientation
b constrained to [a]. alpha remains fixed. beta remains fixed. gamma remains fixed.Peakref> go3
Refining 7 parameters: 199 evaluations in 118 iterations. residue 0.07527 ====
Refining 7 parameters: 99 evaluations in 58 iterations. residue 0.07535
Refining 7 parameters: 69 evaluations in 40 iterations. residue 0.07529
One matrix. 420395 reflections. (forbidden mm:1 weak:43 total:44)
Used: 420351 mm 91845 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.68750 57.69087 -0.00337 57.69090 -0.00340 0.28845
b Con 57.68750 57.69084 0.28845
c Yes 149.69754 149.68208 0.01546 149.68208 0.01546 0.74841
alpha Fix 90.00000 89.99943 1.00000
beta Fix 90.00000 89.99971 1.00000
gamma Fix 90.00000 89.99989 1.00000
orx Yes 0.60693 0.60687 0.00006 0.60687 0.00006 0.00426
ory Yes -0.67102 -0.67112 0.00010 -0.67112 0.00010 0.00426
ora Yes 173.03310 173.04532 -0.01223 173.04532 -0.01223 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes 0.06011 0.06692 -0.00681 0.00000 0.06011 0.10000
zerover Yes 0.06229 0.05692 0.00537 0.00000 0.06229 0.10000
detrotx No 0.00000 0.00000 0.20000
detroty No 0.00000 0.00000 0.20000
detrotz No 0.00000 0.00000 0.20000
=====================================================================
Vol 498170.63 498177.31 -6.69 498177.34 -6.72 420351
=====================================================================
mm 0.06801 0.06313 0.00487 0.11092 -0.04291 420351
mmAng + 0.03362 0.03121 0.00241 0.05483 -0.02122 420351
rotpartial + 0.04167 0.04905 -0.00737 0.04906 -0.00739 91845
rotoutside 0.00000 0.00000 0.00000 0.12527 -0.12527 3
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 328503
rotall 0.00911 0.01072 -0.00161 0.01072 -0.00161 420351
res 0.07529 0.08026 -0.00496 0.10389 -0.02860
Allow the detector to rotate
Refining 10 parameters: 182 evaluations in 113 iterations. residue 0.0748 ====
Refining 10 parameters: 134 evaluations in 81 iterations. residue 0.0748 ====
Refining 10 parameters: 82 evaluations in 49 iterations. residue 0.0748
One matrix. 420395 reflections. (forbidden mm:1 weak:43 total:44)
Used: 420351 mm 91845 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.68800 57.68750 0.00049 57.69090 -0.00291 0.28845
b Con 57.68800 57.69084 0.28845
c Yes 149.69516 149.69754 -0.00238 149.68208 0.01308 0.74841
alpha Fix 90.00000 89.99943 1.00000
beta Fix 90.00000 89.99971 1.00000
gamma Fix 90.00000 89.99989 1.00000
orx Yes 0.60695 0.60693 0.00002 0.60687 0.00008 0.00426
ory Yes -0.67102 -0.67102 0.00000 -0.67112 0.00011 0.00426
ora Yes 173.02960 173.03310 -0.00350 173.04532 -0.01572 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist No 0.00000 0.00000 0.10000
zerohor Yes 0.06053 0.06011 0.00042 0.00000 0.06053 0.10000
zerover Yes 0.06670 0.06229 0.00441 0.00000 0.06670 0.10000
detrotx Yes 0.00890 0.00000 0.00890 0.00000 0.00890 0.20000
detroty Yes -0.00654 0.00000 -0.00654 0.00000 -0.00654 0.20000
detrotz Yes -0.00055 0.00000 -0.00055 0.00000 -0.00055 0.20000
=====================================================================
Vol 498171.31 498170.63 0.69 498177.34 -6.03 420351
=====================================================================
mm 0.06740 0.06801 -0.00061 0.11092 -0.04352 420351
mmAng + 0.03332 0.03362 -0.00030 0.05483 -0.02152 420351
rotpartial + 0.04148 0.04167 -0.00019 0.04906 -0.00758 91845
rotoutside 0.00000 0.00000 0.00000 0.12527 -0.12527 3
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 328503
rotall 0.00906 0.00911 -0.00004 0.01072 -0.00166 420351
res 0.07480 0.07529 -0.00049 0.10389 -0.02909
And also the detector distance
Refining 11 parameters: 226 evaluations in 140 iterations. residue 0.07535
Refining 11 parameters: 187 evaluations in 119 iterations. residue 0.07518
Refining 11 parameters: 114 evaluations in 71 iterations. residue 0.07512
One matrix. 420395 reflections. (forbidden mm:1 weak:43 total:44)
Used: 420351 mm 91845 rot
ref current previous change initial change shift
=====================================================================
a Yes 57.68429 57.68800 -0.00370 57.69090 -0.00661 0.28845
b Con 57.68429 57.69084 0.28845
c Yes 149.68896 149.69516 -0.00620 149.68208 0.00688 0.74841
alpha Fix 90.00000 89.99943 1.00000
beta Fix 90.00000 89.99971 1.00000
gamma Fix 90.00000 89.99989 1.00000
orx Yes 0.60693 0.60695 -0.00002 0.60687 0.00006 0.00426
ory Yes -0.67102 -0.67102 -0.00001 -0.67112 0.00010 0.00426
ora Yes 173.03036 173.02960 0.00076 173.04532 -0.01496 1.00000
xtalx No 0.00000 0.00000 0.10000
xtaly No 0.00000 0.00000 0.10000
xtalz Fix 0.00000 0.00000 0.10000
zerodist Yes -0.00999 0.00000 -0.00999 0.00000 -0.00999 0.10000
zerohor Yes 0.05432 0.06053 -0.00621 0.00000 0.05432 0.10000
zerover Yes 0.06026 0.06670 -0.00644 0.00000 0.06026 0.10000
detrotx Yes 0.00696 0.00890 -0.00194 0.00000 0.00696 0.20000
detroty Yes -0.00445 -0.00654 0.00209 0.00000 -0.00445 0.20000
detrotz Yes -0.00256 -0.00055 -0.00202 0.00000 -0.00256 0.20000
=====================================================================
Vol 498086.63 498171.31 -84.69 498177.34 -90.72 420351
=====================================================================
mm 0.06754 0.06740 0.00014 0.11092 -0.04339 420351
mmAng + 0.03339 0.03332 0.00007 0.05483 -0.02145 420351
rotpartial + 0.04173 0.04148 0.00025 0.04906 -0.00733 91845
rotoutside 0.00000 0.00000 0.00000 0.12527 -0.12527 3
rotinside 0.00000 0.00000 0.00000 0.00000 0.00000 328503
rotall 0.00912 0.00906 0.00005 0.01072 -0.00160 420351
res 0.07512 0.07480 0.00032 0.10389 -0.02878
Now compare the original and refined cells
a b c alpha beta gamma volume
Refined 1 : 57.684 57.684 149.689 90.00 90.00 90.00 498086.6
Original 1 : 57.691 57.691 149.682 90.00 90.00 90.00 498177.4
Volume ratio = 1.0 Trying 32 solutions
Nr Rotangle Rotvec(xyz) RotVec(hkl) ( angle) RotVec(uvw) ( angle) Fom
1 179.983 -0.4336 0.6430 0.6313 0.00 -0.00 -1.00 ( 0.00) 0.00 -0.00 -1.00 ( 0.00) 0.000
2 179.988 -0.8632 -0.0953 -0.4958 -0.00 1.00 -0.00 ( 0.02) -0.00 1.00 -0.00 ( 0.02) 0.000
3 179.992 -0.4274 0.4700 -0.7723 -1.00 1.00 -0.00 ( 0.02) -1.00 1.00 -0.00 ( 0.02) 0.000
4 89.983 -0.4335 0.6430 0.6314 0.00 -0.00 -1.00 ( 0.00) 0.00 -0.00 -1.00 ( 0.00) 0.000
5 90.017 0.4337 -0.6430 -0.6313 -0.00 -0.00 1.00 ( 0.00) -0.00 -0.00 1.00 ( 0.00) 0.000
6 179.991 -0.7933 -0.6047 0.0710 1.00 1.00 0.00 ( 0.00) 1.00 1.00 0.00 ( 0.02) 0.000
7 179.999 -0.2587 -0.7599 0.5963 1.00 0.00 0.00 ( 0.00) 1.00 0.00 0.00 ( 0.00) 0.000
8 0.021 0.8634 -0.4286 -0.2662 -0.11 -1.08 4.00 ( 3.81) -1.97-20.00 11.01 ( 0.06) 0.000
The original and refined cells differ a little bit. The final orientation
can be expressed as a 0.02 degree rotation around an arbitrary axis.
Save all results. The changes are small, so this is not actually
necessary, but these files could be used for reprocessing all the images.
Peakref>
saveall
Adding original position(0.09686 -0.02563 -0.51421) to refined detzero. Adding original angles (0.14892 0.27797 0.35661) to refined detrot.Enter name [detalign.vic] detalign.vic
WARNING: renaming existing file detalign.vic into detalign.vic.~3~ Created detalign.vicEnter name [goniostat.vic] goniostat.vic
WARNING: renaming existing file goniostat.vic into goniostat.vic.~1~
Created goniostat.vic
initial new
-0.0044853 -0.0149614 0.0028971 -0.0044829 -0.0149643 0.0028967
-0.0131739 -0.0016492 -0.0042951 -0.0131726 -0.0016536 -0.0042960
0.0103339 -0.0085963 -0.0042181 0.0103399 -0.0085944 -0.0042170
Determinant: 0.2007317E-05 0.2007683E-05
SameRmat=false
Enter name [icr]
icr.rmat
Created icr.rmat
Fix all but the cell parameters, end determine sigma's with two
methods
Peakref>
fix all
Peakref>
free cell
b constrained to [a]. alpha remains fixed. beta remains fixed. gamma remains fixed.Method 1: apply some errors to the observed positions and refine. Repeat this procedure 50 times.
Calculating sigma's using sigma.
50 cycles of refinement with artificial errors on observed impact positions: f*0.06754 in mm, g*0.04173 rotation in degrees
(f and g a random number between -0.5 and 0.5).
res=0.07511 nref(mm)=420351
a c Volume
refined 57.68429 149.68896 498086.625
sigma 0.00033 0.00361 6.428
420351 reflections. Resolution(d) from 45.69169 to 1.6 Angstrom. Theta from 0.93299 to 27.71021 degrees.
Method 2. Refine parameters against a 10% random subset of the
reflections. Repeat this procedure 50 times.
Calculating sigma's using sigrnd.
50 cycles of refinement using a random subset (fraction=0.1 n=42039) of all 420395 reflections.
res=0.07511 nref(mm)=420351
a c Volume
refined 57.68429 149.68896 498086.625
sigma 0.00026 0.00299 7.000
420351 reflections. Resolution(d) from 45.69169 to 1.6 Angstrom. Theta from 0.93299 to 27.71021 degrees.
Peakref>
exit
peakref ended at 25-Nov-2010 21:47:10 CPU time used 00:13:29
any (version 1.5 2010111000) started on bekken at 25-Nov-2010 21:47:10 Appending to any.logAny> read final
y-data from final.y
465397 reflections nPg=8 (h,k,l) (k,-h,l) (-h,-k,l) (-k,h,l) (-h,-k,-l) (-k,h,-l) (h,k,-l) (k,-h,-l)
Start mdian2 on 465397 reflections mdian2 stopped after 101 passes median=31956.0 presentationscale=0.0000626
Calculating Extremes
Cannot calculate icr for reflection 54. (total 1853 icr's failed)
69 too negative reflections (allowed) 34626 weak reflections (allowed)
Status for final.y
RMAT 1
RMAT DMAT
-0.0044853 -0.0149614 0.0028971 -14.9281244 -43.8460999 34.3934746
-0.0131739 -0.0016492 -0.0042951 -49.7948074 -5.4893670 -28.6107864
0.0103339 -0.0085963 -0.0042181 64.9072723 -96.2312393 -94.5056610
Determinant: 0.2007317E-05 498177.4
Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/m
Lambda=1.488 pointgroup=4/m
reflections=465397 unique=64247 weak=34626 negative=69
Unique: H from -36 to 0 K from -25 to 24 L from -93 to 0
Full Sphere: H from -36 to 36 K from -36 to 36 L from -93 to 93
Max equivalents 11 datalimit theta 0.8544 27.7102. datalimit reso 1.6 49.894. datalimit chi 0.0 78.1585.
datalimit psi -153.2682 153.0809.
datalimit intensity -108.0624 16449.5723. datalimit duration 1.0 5.0.
one Experiment 368 Sets 369 Images
presentationscale 0.0000626
Selectable: GOOD WEAK EDGEVER EDGEHOR EDGEROT BIGHOR NEGATIVE BADUNIF OVERFLOW
Forbid: EDGEVER EDGEHOR EDGEROT BIGHOR BADUNIF OVERFLOW
Allow: GOOD WEAK NEGATIVE
Require: NONE
Rsym=0.097 Rmeas=0.105 Rpim=0.039 Chi2=28.511 nRsym=453087 Unique1=45 Unique2+=64113
<I>=194.609 <s>=4.871 <I/s>=27.387 <I>/<s>=39.95 nMean=453132
Create a summary of reflections 'flags'
Ecmaxflag=23 maxcode=16777215
--------------------------------------------------------------------------------
Flags for all reflections
Good 419896 Not Good 45501 (Weak 34626 Not Weak 10875) Total: 465397
Total Percent NonWeak Percent
419896 90.223 419896 97.475 GOOD
34626 7.440 WEAK
373 0.080 240 0.056 EDGEVER
44 0.009 33 0.008 EDGEHOR
337 0.072 294 0.068 EDGEROT
27 0.006 16 0.004 BIGHOR
69 0.015 69 0.016 NEGATIVE
9595 2.062 BADUNIF
2341 0.503 2340 0.543 OVERFLOW
Get a table of intensity statistics.
Any>
list intensity
-------------------------------------------------------------------------------- Intensity distribution for Shells Forbid: EDGEVER EDGEHOR EDGEROT BIGHOR BADUNIF OVERFLOW Allow: GOOD WEAK NEGATIVE Require: NONE Sh Theta Reso n1 <I> <sig> <I>/<sig> <I/sig> 1 12.46 3.45 47677 668.58 13.70 48.82 45.01 2 15.78 2.74 47180 427.52 9.08 47.07 41.12 3 18.14 2.39 47538 213.15 4.94 43.14 35.74 4 20.04 2.17 47429 174.02 4.28 40.66 33.18 5 21.66 2.02 46840 133.71 3.60 37.13 29.25 6 23.09 1.90 46124 95.88 2.97 32.28 25.35 7 24.39 1.80 45920 60.02 2.39 25.09 19.69 8 25.57 1.72 45886 43.86 2.22 19.78 15.95 9 26.68 1.66 44909 32.52 2.16 15.07 12.35 10 27.71 1.60 33629 24.88 2.19 11.34 9.54 =================================================== 27.71 1.60 453132 194.58 4.87 39.94 27.39
Save the reflections for sadabs. Include the too-strong reflections.
Any>
allow overflow
Forbid: EDGEVER EDGEHOR EDGEROT BIGHOR BADUNIF Allow: GOOD WEAK NEGATIVE OVERFLOW Require: NONE
Sadabs will establish an error model for the dataset. So do not modify
the sigma's of the intensities. A good value for msa will be calculated
by Sadabs. In principle I would use msa 0.0, but a bug in sadabs will
reject reflections with I/sigma > 1000, so use a small value for now.
Any>
msa 0.001
69 too negative reflections (allowed) 34619 weak reflections (allowed)Any> sadabs
msa = 0.001. Sadabs likes to have it zero. Continuing... Created shelx.sad Created shelx.par Created shelx.any cell 1 written as target with id 1 Rmat ok minI -262822.0 maxI 262831264.0 minSigI 486.0 Maximum intensity (262831264.0) scaled to 9999999.0 with scale factor 0.03805 455469 lines written.Any> exit
Closed shelx.sad any ended at 25-Nov-2010 21:48:07 CPU time used 00:00:53ic> cd -
SADABS-2008/1 - Bruker AXS area detector scaling and absorption correction -------------------------------------------------------------------------- Note that all questions except those asking for a filename etc. may be answered by "Q" to force SADABS to terminate immediately.Expert mode (Y or N) [N]: n
Laue group numbers: [1] -1 [8] -3m (rhombohedral axes) [2] 2/m (Y unique) [9] -31m (Z unique) [3] mmm [10] -3m1 (Z unique) [4] 4/m (Z unique) [11] 6/m (Z unique) [5] 4/mmm (Z unique) [12] 6/mmm (Z unique) [6] -3 (rhombohedral axes) [13] m3 [7] -3 (Z unique) [14] m3m [0] to write list of equivalent indices for Laue/point groups to listing fileEnter Laue group number [2]: 5
Equivalent reflections defined by point group 4/mmm for scaling Equivalent reflections defined by point group 4/mmm for error model Read reflection files written by EVALCCD (with extension .sad specified) or by SAINT (extension .raw, default if no extension, or .ram for incommensurate structures). It is important that all files are from the same crystal and that reflections have been indexed consistently, i.e. that the orientation matrices are similar (no rows with signs reversed)! Note that XPREP can reindex a .raw or .sad file transforming the direction cosines. It is also possible to read in a .mul file from the integration of a twinned crystal and select just one twin domain for processing using the SAINT partitioning.Enter filename (/ if no more) [ ]: shelx.sad
Mean and maximum errors in direction cosine check function = 0.000 0.000 The mean error should not exceed 0.005, and is usually caused by matrix changes during data processing. Approximate wavelength, cell and maximum 2-theta (from cosines etc.): 1.48826 57.702 57.702 149.710 89.999 90.000 90.000 55.50 ============================================================================== PART 1 - Refinement of parameters to model systematic errorsRestraint esd for equal consecutive scale factors [0.020]: 0.005
Weak (W), moderate (M) or strong (S) empirical corrections or face-indexednumerical absorption correction (N) [M]: m
434653 Reflections employed for parameter determination
Effective data to parameter ratio = 983.02
wR2(int) = 0.1238 (selected reflections only, before parameter refinement)
Cycle wR2(incid) wR2(diffr) Mean wt.
1 0.1117 0.1014 0.8228
2 0.0985 0.0959 0.8462
3 0.0943 0.0931 0.8558
4 0.0920 0.0913 0.8615
5 0.0905 0.0902 0.8653
6 0.0895 0.0894 0.8678
7 0.0889 0.0889 0.8695
8 0.0885 0.0886 0.8706
9 0.0882 0.0884 0.8714
10 0.0880 0.0882 0.8719
11 0.0879 0.0881 0.8723
12 0.0878 0.0881 0.8726
13 0.0878 0.0880 0.8728
14 0.0877 0.0880 0.8729
15 0.0877 0.0880 0.8730
16 0.0877 0.0879 0.8731
17 0.0877 0.0879 0.8732
18 0.0877 0.0879 0.8732
19 0.0877 0.0879 0.8733
20 0.0877 0.0879 0.8733
21 0.0877 0.0879 0.8733
22 0.0877 0.0879 0.8733
23 0.0877 0.0879 0.8733
24 0.0877 0.0879 0.8734
25 0.0876 0.0879 0.8734
wR2(int) = 0.0879 (selected reflections only, after parameter refinement)
Repeat parameter refinement (R) or accept (A) [A]:
a
============================================================================== PART 2 - Reject outliers and establish error model Rejected reflections are ignored in the statistics and Postscript plots (except the detector diagnostics) and in the output .hkl file Before applying rejections there are: 455469 total and 34400 unique reflections assuming Friedel's law. 426658 total and 34396 unique reflections left after |I-<I>|/su test g = 0.0462 gives best error model.Enter new value for g or <CR> to accept: <Enter>
Run 2-theta R(int) Incid. factors Diffr. factors K Total I>2sig(I)
1 0.0 0.0549 0.602 - 1.033 0.805 - 1.303 1.518 426658 393438
su = K * Sqrt[ sigma^2(I) + (g<I>)^2.00 ] where sigma(I) is from EVALCCD
The above statistics are based on all non-rejected data, ignoring
reflections without equivalents when estimating R(int) and K.
Repeat parameter refinement (P) or accept (A) [A]:
a
============================================================================== PART 3 - Output Postscript diagnostics and corrected dataWrite Postscript diagnostic file (Y or N) [Y]: y
Repeat (R), write unmerged .hkl (W), merged .hkl (M), .sca (S), XD format (D),modulated .hk6 (J), testxtl.dat (BioXhit) (T) or quit (Q) [W]: w
426658 Corrected reflections written to file sad.hkl Estimated minimum and maximum transmission: 0.4597 0.7456 The ratio of these values is more reliable than their absolute values! Repeat (R), write unmerged .hkl (W), merged .hkl (M), .sca (S), XD format (D),modulated .hk6 (J), testxtl.dat (BioXhit) (T) or quit (Q) [Q]: q
name=shellscript-3
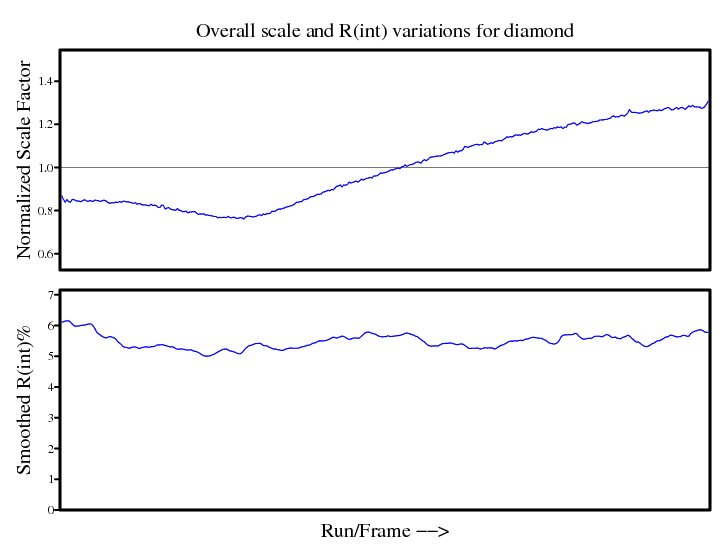
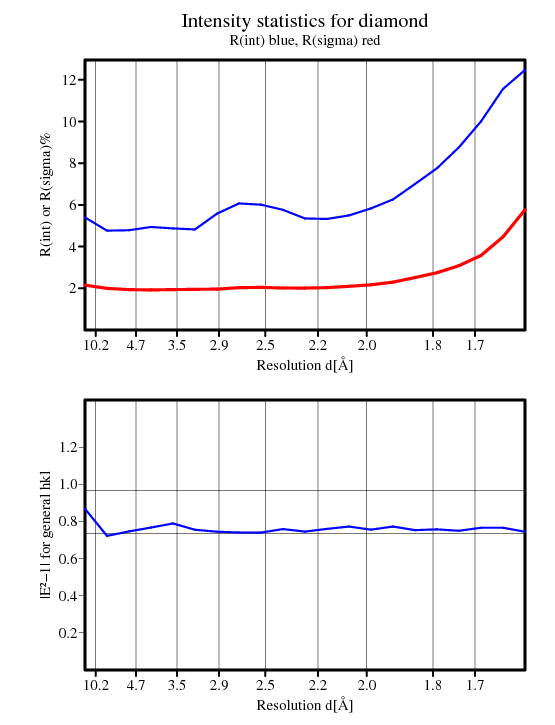
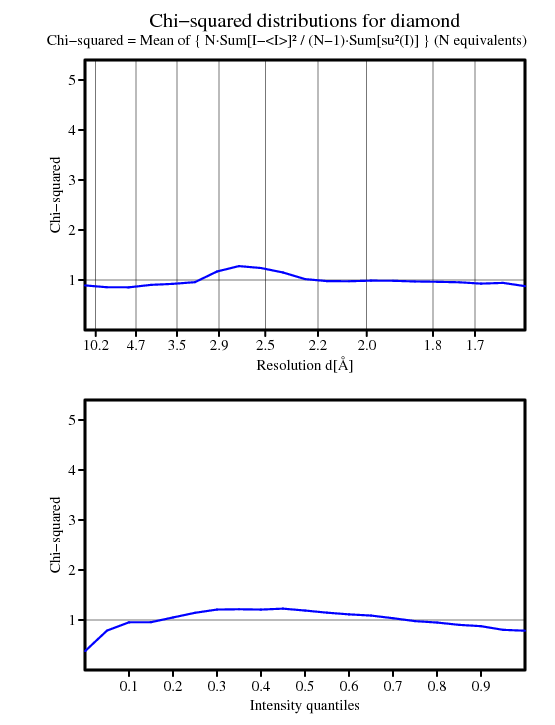
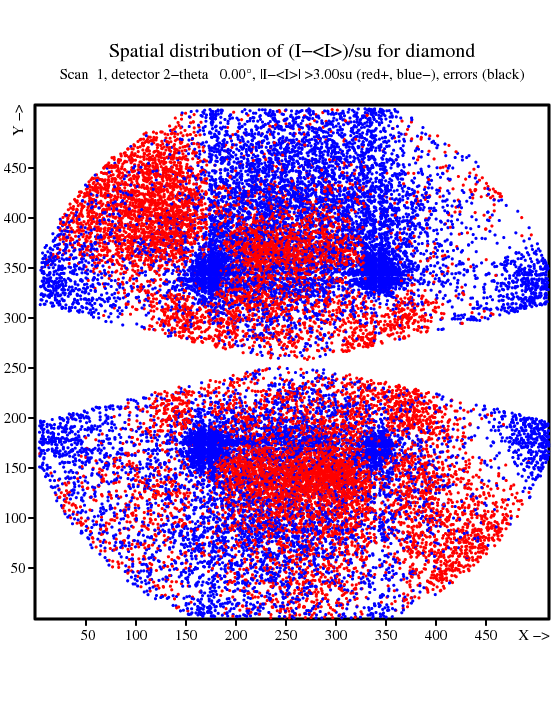
any (version 1.5 2010111000) started on bekken at 25-Nov-2010 21:48:46 Appending to any.logAnd now some statistics
Read the resulting reflection file created by SadAbs.
First read a small eval15 output file. This will initialize
some parameters (wavelength, cell parameters.....)
Any>
read s01f001
y-data from s01f001.y
1264 reflections nPg=8 (h,k,l) (k,-h,l) (-h,-k,l) (-k,h,l) (-h,-k,-l) (-k,h,-l) (h,k,-l) (k,-h,-l)
Start mdian2 on 1264 reflections median=32160.002 presentationscale=0.0000622
Calculating Extremes
Cannot calculate icr for reflection 1. (total 223 icr's failed)
102 weak reflections (allowed)
Status for s01f001.y
RMAT 1
RMAT DMAT
-0.0044853 -0.0149614 0.0028971 -14.9281244 -43.8460999 34.3934746
-0.0131739 -0.0016492 -0.0042951 -49.7948074 -5.4893670 -28.6107864
0.0103339 -0.0085963 -0.0042181 64.9072723 -96.2312393 -94.5056610
Determinant: 0.2007317E-05 498177.4
Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/m
Lambda=1.488 pointgroup=4/m
reflections=1264 unique=1261 weak=102
Unique: H from -35 to 0 K from -25 to 24 L from -93 to 0
Full Sphere: H from -36 to 36 K from -36 to 36 L from -93 to 93
Max equivalents 2 datalimit theta 2.9599 27.7016. datalimit reso 1.6005 14.4081. datalimit chi 0.0364 71.9695.
datalimit psi -144.8713 149.0528.
datalimit intensity -2.0244 5594.459. datalimit duration 1.004 4.998.
one Experiment one Set one Image
presentationscale 0.0000622
Input Flags: GOOD WEAK EDGEVER EDGEROT NEGATIVE BADUNIF OVERFLOW
Selectable: GOOD WEAK EDGEVER EDGEROT BADUNIF OVERFLOW
Forbid: EDGEVER EDGEROT BADUNIF OVERFLOW
Allow: GOOD WEAK
Require: NONE
Rsym=0.034 Rmeas=0.048 Rpim=0.034 Chi2=2.002 nRsym=4 Unique1=1042 Unique2+=2
<I>=201.849 <s>=4.994 <I/s>=27.753 <I>/<s>=40.421 nMean=1046
and set the pointgroup
pointgroup set to 4/mmm Calculating Extremes Cannot calculate icr for reflection 1. (total 222 icr's failed) nPg=16 (h,k,l) (k,-h,l) (-h,-k,l) (-k,h,l) (h,-k,-l) (-k,-h,-l) (-h,k,-l) (k,h,-l) (-h,-k,-l) (-k,h,-l) (h,k,-l) (k,-h,-l) (-h,k,l) (k,h,l) (h,-k,l) (-k,-h,l)Then read the actual scaled hkl file:
Using current rmat and wavelength Lambda=1.488
RMAT 1
RMAT DMAT
-0.0044853 -0.0149614 0.0028971 -14.9281244 -43.8460999 34.3934746
-0.0131739 -0.0016492 -0.0042951 -49.7948074 -5.4893670 -28.6107864
0.0103339 -0.0085963 -0.0042181 64.9072723 -96.2312393 -94.5056610
Determinant: 0.2007317E-05 498177.4
Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/mmm
hkl-data from sad.hkl
Opened sad.hkl 426658 reflections
Start mdian2 on 426658 reflections median=3.591 presentationscale=0.5568936
Calculating Extremes
Cannot calculate icr for reflection 760. (total 805 icr's failed)
38085 weak reflections (allowed)
Status for sad.hkl
RMAT 1
RMAT DMAT
-0.0044853 -0.0149614 0.0028971 -14.9281244 -43.8460999 34.3934746
-0.0131739 -0.0016492 -0.0042951 -49.7948074 -5.4893670 -28.6107864
0.0103339 -0.0085963 -0.0042181 64.9072723 -96.2312393 -94.5056610
Determinant: 0.2007317E-05 498177.4
Cell: 57.69090 57.69084 149.68208 89.9994 89.9997 89.9999 V= 498177.41
Sigma 0.0408 0.0209 0.0406 0.014 0.014 0.035 Volume 158.84
Bravais=P pg=4/mmm
Lambda=1.488 pointgroup=4/mmm
reflections=426658 unique=34396 weak=38085
Unique: H from -36 to 0 K from -25 to 0 L from -93 to 0
Full Sphere: H from -36 to 36 K from -36 to 36 L from -93 to 93
Max equivalents 19 datalimit theta 0.8544 27.7102. datalimit reso 1.6 49.894.
datalimit intensity -2.2917 5568.9312.
one Experiment one Set
presentationscale 0.5568936
Input Flags: GOOD WEAK NEGATIVE
Selectable: GOOD WEAK
Forbid: NEGATIVE
Allow: ALL
Require: NONE
Rsym=0.055 Rmeas=0.057 Rpim=0.016 Chi2=1.012 nRsym=426647 Unique1=11 Unique2+=34385
<I>=72.161 <s>=5.412 <I/s>=10.236 <I>/<s>=13.333 nMean=426658
Intensity statistics
-------------------------------------------------------------------------------- Intensity distribution for Shells Forbid: NEGATIVE Allow: ALL Require: NONE Sh Theta Reso n1 <I> <sig> <I>/<sig> <I/sig> 1 12.46 3.45 47046 260.51 18.35 14.20 13.79 2 15.78 2.74 42854 154.05 10.94 14.08 13.27 3 18.14 2.39 41557 77.22 5.60 13.78 12.40 4 20.04 2.17 44585 62.39 4.61 13.54 12.00 5 21.66 2.02 44412 47.78 3.64 13.13 11.18 6 23.09 1.90 43879 33.88 2.73 12.43 10.31 7 24.39 1.80 43949 20.94 1.91 10.98 8.78 8 25.57 1.72 43821 15.11 1.59 9.51 7.60 9 26.68 1.66 42760 11.13 1.42 7.85 6.29 10 27.71 1.60 31795 8.57 1.37 6.24 5.10 =================================================== 27.71 1.60 426658 72.15 5.41 13.33 10.24Diffraction up to above 1.6 Å
Completeness and redundancy
Any>
rshell
-------------------------------------------------------------------------------- Completeness and Rmerge for Shells Forbid: NEGATIVE Allow: ALL Require: NONE Sh Theta Reso Meas Equi Obs Mis Total Perc Cum Uni1 Uni2+ Nrsym Redun Rsym Rmeas Rpim Chi2 1 12.46 3.45 43697 7365 51062 20 51082 100.0 100.0 3 3748 47043 12.55 0.049 0.051 0.014 0.89 2 15.78 2.74 38703 12023 50726 0 50726 100.0 100.0 2 3491 42852 12.27 0.052 0.054 0.015 1.10 3 18.14 2.39 36825 14085 50910 2 50912 100.0 100.0 2 3452 41555 12.04 0.060 0.062 0.018 1.25 4 20.04 2.17 38628 12468 51096 16 51112 100.0 100.0 1 3434 44584 12.98 0.054 0.056 0.015 1.05 5 21.66 2.02 37945 13043 50988 0 50988 100.0 100.0 0 3411 44412 13.02 0.055 0.057 0.016 0.99 6 23.09 1.90 37019 13689 50708 0 50708 100.0 100.0 0 3374 43879 13.01 0.060 0.062 0.017 1.00 7 24.39 1.80 36685 14209 50894 0 50894 100.0 100.0 0 3377 43949 13.01 0.070 0.073 0.020 0.98 8 25.57 1.72 36253 15041 51294 0 51294 100.0 100.0 0 3394 43821 12.91 0.082 0.086 0.024 0.97 9 26.68 1.66 35099 15603 50702 0 50702 100.0 100.0 0 3347 42762 12.78 0.099 0.103 0.029 0.95 10 27.71 1.60 25816 25114 50930 8 50938 100.0 100.0 3 3356 31788 9.47 0.121 0.128 0.041 0.94 ======================================================================================================== 27.71 1.60 366670 142640 509310 46 509356 100.0 100.0 11 34384 426645 12.41 0.055 0.057 0.016 1.01
any ended at 25-Nov-2010 21:48:52 CPU time used 00:00:06ic> cd -