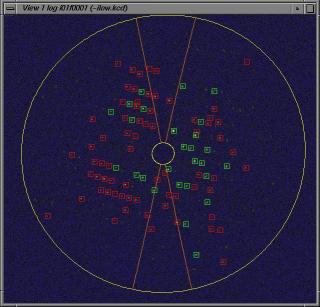
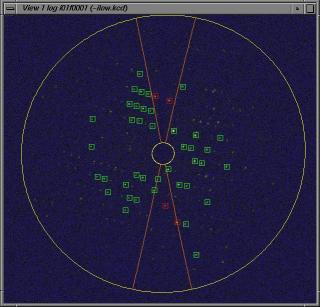
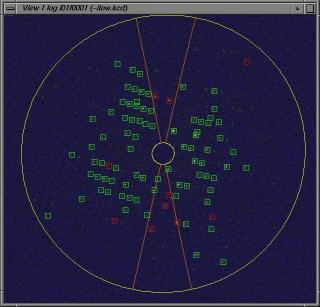
View phi/chi Cell Determination
The Phi/Chi procedure is described in
- A. J. M. Duisenberg, R. W. W. Hooft,
A. M. M. Schreurs and J. Kroon.
Accurate cells from area-detector images.
J. Appl. Cryst. 33 (2000) 893-898
If the default cell determination fails you could try to find the
unit cell using the program view. This document gives an example of the
procedure you could follow.
Suppose you have tried a phichi datacollection
resulting in 8 images i01f0001.kcd
to i08f0001.kcd.
To get rid of systematics in the background, you create a
background image using
backgroundimage i0?f001.kcd write=ilow.kcd.
This image can be subtracted from all the images during peak search.
If there is no file ilow.kcd available,
the peaksearch will still be possible.
2 scripts (i.vic and phichi.vic)
are needed for the peaksearch. The program
buildphichi
will create these files.
phichi.vic
! abort if not called with 2 parameters
\if not alias $1' \exit
\if not alias $2' \exit
! example: $1=i01f $2=i02f
! read and plot the phi scan i01f0001.kcd
readwait '$1'0001 plot
! Calculate theta on the centres of the four
! edges of the detector. thmax will be set to
! the maximum value of these four numbers.
thhalf
! set peakfile name to i01f.pk
peakfile '$1'
! create peakfile, do peaksearch and close peakfile
peakopen peak2 peakclose
! create second window
activate 2
! read and plot phichi scan i02f0001.kcd
readwait '$2'0001 plot
! plot chiarcs from i01f.pk
peakchifile '$1'
! set peakfile name to i02f.pk
peakfile '$2'
! create peakfile, do peaksearch and close peakfile
peakopen peak2 peakclose
! leave script with first window active
activate 1
This script is used to search peakcoordinates in the phi and phichi
images. The peaks will be stored in files
i01f.pk, i02f.pk, .....
Needed:
inspection of i01f0001.kcd
i.vic
! trigger fatal errors instead of warnings
abort on
! the standard detalign.vic will be used.
! a detalign.vic in the current directory will
! overrule the standard one. If you created a
! detalign_i.vic, use it.
\if file detalign_i.vic @detalign_i
! set the number of expected peaks in one image
peaknr 80
! the peaksurrounding box.
peaksizemm 2.2
! stop peak search after 5 successive weak peaks
peakbreak 5
! set minimum treshhold. No peaks will be searched
! with a maximum pixel value less than this treshhold.
! a good default (if you use ilow.kcd) is 10
peakmin 5
! set theta min. theta max will be set in phichi.vic
thmin 3.0
! define region with too long active reflections
durationmax 5
! each image will be scaled to the maximum pixel value
noglobalmax
! display peak numbers
peaklabel on
! set default colour scheme
mode jaap
! if ilow.kcd exists, use it and change colour scheme
! compressed files (ilow.kcd.Z ilow.kcd.gz ilow.kcd.bz2)
! are also permitted.
\if not filez ilow.kcd \goto skipdark
dark ilow.kcd
mode log
!skipdark
! do the peak search. Use phichi.vic with two parameters
! 1 = filename of phi scan (assumed frame number 0001)
! 2 = filename of phichi scan (assumed frame number 0001)
@phichi(i01f;i02f)
@phichi(i03f;i04f)
@phichi(i05f;i06f)
@phichi(i07f;i08f)
Do the peaksearch with the command:
view @i.
This will result the following 8 new files:
i01f.pk to i08f.pk
run the upc program.
upc i 0.156 10.0 0.05 0.01
These are the default arguments.
| i |
name of pk files |
| 0.156 |
radius margin |
| 10.0 |
sigma factor intensity differences |
| 0.05 |
edge fraction |
| 0.01 |
Maximum c-vector difference for Friedel merge |
peaks from the phi and phichi images have to be combined. Two numbers
are used:
- radius
This is the distance on the detector from the
impact of the primary beam to the impact of the peak. This radius
should be equal (within margins) for corresponding peaks. The default
margin is calculated from the pixelsize in the pkfile.
- sigma factor
Intensities of corresponding peaks should be similar.
The sigma of the intensities of a possible pair is calculated as
sigma = sqrt( 0.5 * ( Iphi + Iphichi ) )
diff = abs( 0.5 * ( Iphi - Iphichi ) )
the pair is allowed if:
diff < factor*sigma
After pairing two peaks, the rotation value will be calculated.
If the resulting angle is within edge fraction of the edge of
the possible range, the resulting reflection will not be used.
After the first part of the upc program 5 new files have been created:
i01f.drx,
i03f.drx,
i05f.drx,
i07f.drx and i.pk
The drx files contain the c-vectors. You may use them to
run dirax manually, but the combined drx file (see below)
will give better results.
The pk file contains impact coordinates and rotation angles. This
file will be used in the refinement program.
The second part of the program will merge Friedel related
reflections. Two number are used:
- sigma factor
Intensities of corresponding peaks should be similar.
The sigma of the intensities of a possible friedel pair is calculated as
sigma = sqrt( 0.5 * ( I1 + I2 ) )
diff = abs( 0.5 * ( I1 - I2 ) )
the reflections will be merged if:
diff < factor*sigma
- Maximum c-vector difference
Two c-vectors will be merged if the difference in their lengths
is less than this fixed maximum difference.
The reduced set of reflections is written to i.drx.
This file
is the input file for the indexing program DirAx.
(The reflections without a Friedel are stored in isingle.drx.)
The upc program will write information to the file
i_upc.lis
run the DirAx program.
dirax i go
Useful options are:
| help |
|
how to use DirAx |
| levelfit |
1000 |
increase for more strict or decrease for more tolerant |
| indexfit |
2 |
decrease for more strict or increase for more tolerant |
| store |
|
save solution |
| lchi |
|
invert fitting/nonfitting reflections |
| go |
|
another go with fitting reflections |
| compare |
|
compare 2 solutions |
| restore |
|
restore a saved solution |
| ccd |
|
save solution |
Start refinement program, read the cell parameters from
i.rmat and the peak positions from
i.pk:
peakref rmat i pk i
| help |
how to use peakref |
| go3 |
3 cycles of refinement |
| free rmat |
allow cell parameters fo refine |
| save |
create detector alignment file detalign.vic |
| savermat |
write refined cell parameters to file |
View Main Page
View Peak Search Page